Influential factors of saliva microbiota composition
- PMID: 36344584
- PMCID: PMC9640688
- DOI: 10.1038/s41598-022-23266-x
Influential factors of saliva microbiota composition
Abstract
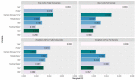
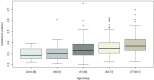
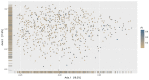
The oral microbiota is emerging as an influential factor of host physiology and disease state. Factors influencing oral microbiota composition have not been well characterised. In particular, there is a lack of population-based studies. We undertook a large hypothesis-free study of the saliva microbiota, considering potential influential factors of host health (frailty; diet; periodontal disease), demographics (age; sex; BMI) and sample processing (storage time), in a sample (n = 679) of the TwinsUK cohort of adult twins. Alpha and beta diversity of the saliva microbiota was associated most strongly with frailty (alpha diversity: β = -0.16, Q = 0.003, Observed; β = -0.16, Q = 0.002, Shannon; β = -0.16, Q = 0.003, Simpson; Beta diversity: Q = 0.002, Bray Curtis dissimilarity) and age (alpha diversity: β = 0.15, Q = 0.006, Shannon; β = 0.12, Q = 0.003, Simpson; beta diversity: Q = 0.002, Bray Curtis dissimilarity; Q = 0.032, Weighted UniFrac) in multivariate models including age, frailty, sex, BMI, frailty and diet, and adjustment for multiple testing. Those with a more advanced age were more likely to be dissimilar in the saliva microbiota composition than younger participants (P = 5.125e-06, ANOVA). In subsample analyses, including consideration of periodontal disease (total n = 138, periodontal disease n = 66), the association with frailty remained for alpha diversity (Q = 0.002, Observed ASVs; Q = 0.04 Shannon Index), but not beta diversity, whilst age was not demonstrated to associate with alpha or beta diversity in this subsample, potentially due to insufficient statistical power. Length of time that samples were stored prior to sequencing was associated with beta diversity (Q = 0.002, Bray Curtis dissimilarity). Six bacterial taxa were associated with age after adjustment for frailty and diet. Of the factors studied, frailty and age emerged as the most influential with regards to saliva microbiota composition. Whilst age and frailty are correlates, the associations were independent of each other, giving precedence to both biological and chronological ageing as processes of potential importance when considering saliva microbiota composition.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

