Prime editing for precise and highly versatile genome manipulation
- PMID: 36344749
- PMCID: PMC10989687
- DOI: 10.1038/s41576-022-00541-1
Prime editing for precise and highly versatile genome manipulation
Abstract
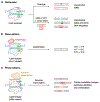
Programmable gene-editing tools have transformed the life sciences and have shown potential for the treatment of genetic disease. Among the CRISPR-Cas technologies that can currently make targeted DNA changes in mammalian cells, prime editors offer an unusual combination of versatility, specificity and precision. Prime editors do not require double-strand DNA breaks and can make virtually any substitution, small insertion and small deletion within the DNA of living cells. Prime editing minimally requires a programmable nickase fused to a polymerase enzyme, and an extended guide RNA that both specifies the target site and templates the desired genome edit. In this Review, we summarize prime editing strategies to generate programmed genomic changes, highlight their limitations and recent developments that circumvent some of these bottlenecks, and discuss applications and future directions.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
The authors have filed patent applications on gene editing technologies through the Broad Institute of MIT and Harvard. P.J.C. is currently an employee of Prime Medicine. D.R.L. is a consultant and equity owner of Beam Therapeutics, Pairwise Plants, Prime Medicine, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering technologies.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

