A gut-derived hormone suppresses sugar appetite and regulates food choice in Drosophila
- PMID: 36344765
- PMCID: PMC9684077
- DOI: 10.1038/s42255-022-00672-z
A gut-derived hormone suppresses sugar appetite and regulates food choice in Drosophila
Abstract
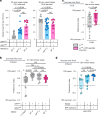
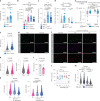
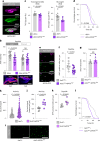
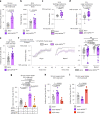
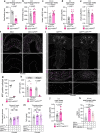
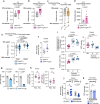
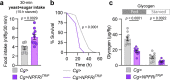
Animals must adapt their dietary choices to meet their nutritional needs. How these needs are detected and translated into nutrient-specific appetites that drive food-choice behaviours is poorly understood. Here we show that enteroendocrine cells of the adult female Drosophila midgut sense nutrients and in response release neuropeptide F (NPF), which is an ortholog of mammalian neuropeptide Y-family gut-brain hormones. Gut-derived NPF acts on glucagon-like adipokinetic hormone (AKH) signalling to induce sugar satiety and increase consumption of protein-rich food, and on adipose tissue to promote storage of ingested nutrients. Suppression of NPF-mediated gut signalling leads to overconsumption of dietary sugar while simultaneously decreasing intake of protein-rich yeast. Furthermore, gut-derived NPF has a female-specific function in promoting consumption of protein-containing food in mated females. Together, our findings suggest that gut NPF-to-AKH signalling modulates specific appetites and regulates food choice to ensure homeostatic consumption of nutrients, providing insight into the hormonal mechanisms that underlie nutrient-specific hungers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Comment in
-
No sugar, just protein please - says the fly.Nat Metab. 2022 Nov;4(11):1436-1437. doi: 10.1038/s42255-022-00665-y. Nat Metab. 2022. PMID: 36344763 No abstract available.
References
-
- Richter CP. Increased salt appetite in adrenalectomized rats. Am. J. Physiol. 1936;115:155–161.
-
- Richter CP. Total self regulatory functions of animals and human beings. Harvey Lect. Ser. 1943;38:63–103.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

