Cryo-EM structures of prefusion SIV envelope trimer
- PMID: 36344847
- PMCID: PMC10606957
- DOI: 10.1038/s41594-022-00852-1
Cryo-EM structures of prefusion SIV envelope trimer
Abstract
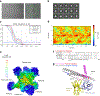
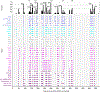
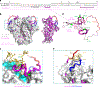
Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion stabilization and cryo-EM structures of soluble envelope (Env) trimers from rhesus macaque SIV (SIVmac) in complex with neutralizing antibodies. These structures provide residue-level definition for SIV-specific disulfide-bonded variable loops (V1 and V2), which we used to delineate variable-loop coverage of the Env trimer. The defined variable loops enabled us to investigate assembled Env-glycan shields throughout SIV, which we found to comprise both N- and O-linked glycans, the latter emanating from V1 inserts, which bound the O-link-specific lectin jacalin. We also investigated in situ SIVmac-Env trimers on virions, determining cryo-electron tomography structures at subnanometer resolutions for an antibody-bound complex and a ligand-free state. Collectively, these structures define the prefusion-closed structure of the SIV-Env trimer and delineate variable-loop and glycan-shielding mechanisms of immune evasion conserved throughout SIV evolution.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

