A haplotype-resolved genome assembly of the Nile rat facilitates exploration of the genetic basis of diabetes
- PMID: 36344967
- PMCID: PMC9641963
- DOI: 10.1186/s12915-022-01427-8
A haplotype-resolved genome assembly of the Nile rat facilitates exploration of the genetic basis of diabetes
Abstract
Background: The Nile rat (Avicanthis niloticus) is an important animal model because of its robust diurnal rhythm, a cone-rich retina, and a propensity to develop diet-induced diabetes without chemical or genetic modifications. A closer similarity to humans in these aspects, compared to the widely used Mus musculus and Rattus norvegicus models, holds the promise of better translation of research findings to the clinic.
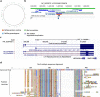
Results: We report a 2.5 Gb, chromosome-level reference genome assembly with fully resolved parental haplotypes, generated with the Vertebrate Genomes Project (VGP). The assembly is highly contiguous, with contig N50 of 11.1 Mb, scaffold N50 of 83 Mb, and 95.2% of the sequence assigned to chromosomes. We used a novel workflow to identify 3613 segmental duplications and quantify duplicated genes. Comparative analyses revealed unique genomic features of the Nile rat, including some that affect genes associated with type 2 diabetes and metabolic dysfunctions. We discuss 14 genes that are heterozygous in the Nile rat or highly diverged from the house mouse.
Conclusions: Our findings reflect the exceptional level of genomic resolution present in this assembly, which will greatly expand the potential of the Nile rat as a model organism.
Keywords: Arvicanthis niloticus; Diabetes; Diurnal; Genome; Germline mutation rate; Heterozygosity; Long-read genome assembly; Orthology; Positive selection; Retrogenes; Segmental duplications.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Yang C, Zhang G, Toh H, et al. Heterozygosity spectrum: all.makeup.agp. Nile rat genome paper supplementary materials on OSF. 2021. 10.17605/OSF.IO/K6EY9.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

