Bacteriophage-encoded lethal membrane disruptors: Advances in understanding and potential applications
- PMID: 36345304
- PMCID: PMC9636201
- DOI: 10.3389/fmicb.2022.1044143
Bacteriophage-encoded lethal membrane disruptors: Advances in understanding and potential applications
Abstract
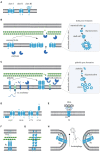
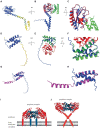
Holins and spanins are bacteriophage-encoded membrane proteins that control bacterial cell lysis in the final stage of the bacteriophage reproductive cycle. Due to their efficient mechanisms for lethal membrane disruption, these proteins are gaining interest in many fields, including the medical, food, biotechnological, and pharmaceutical fields. However, investigating these lethal proteins is challenging due to their toxicity in bacterial expression systems and the resultant low protein yields have hindered their analysis compared to other cell lytic proteins. Therefore, the structural and dynamic properties of holins and spanins in their native environment are not well-understood. In this article we describe recent advances in the classification, purification, and analysis of holin and spanin proteins, which are beginning to overcome the technical barriers to understanding these lethal membrane disrupting proteins, and through this, unlock many potential biotechnological applications.
Keywords: alphaFold structure; bacteriophage; holins spanins; holins spanins applications; holins spanins future potential potential; membrane protein.
Copyright © 2022 Abeysekera, Love, Manners, Billington and Dobson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ahammad T., Drew D. L., Sahu I. D., Serafin R. A., Clowes K. R., Lorigan G. A. (2019). Continuous wave electron paramagnetic resonance spectroscopy reveals the structural topology and dynamic properties of active pinholin S2168 in a lipid bilayer. J. Phys. Chem. B 123, 8048–8056. doi: 10.1021/acs.jpcb.9b06480, PMID: - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

