Altered gene expression in human brain microvascular endothelial cells in response to the infection of influenza H1N1 virus
- PMID: 36345345
- PMCID: PMC9631584
- DOI: 10.1186/s44149-022-00053-9
Altered gene expression in human brain microvascular endothelial cells in response to the infection of influenza H1N1 virus
Abstract
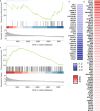
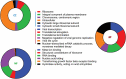
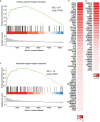
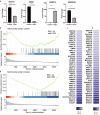
Influenza viruses not only cause respiratory illness, but also have been reported to elicit neurological manifestations following acute viral infection. The central nervous system (CNS) has a specific defense mechanism against pathogens structured by cerebral microvasculature lined with brain endothelial cells to form the blood-brain barrier (BBB). To investigate the response of human brain microvascular endothelial cells (hBMECs) to the Influenza A virus (IAV), we inoculated the cells with the A/WSN/33 (H1N1) virus. We then conducted an RNAseq experiment to determine the changes in gene expression levels and the activated disease pathways following infection. The analysis revealed an effective activation of the innate immune defense by inducing the pattern recognition receptors (PRRs). Along with the production of proinflammatory cytokines, we detected an upregulation of interferons and interferon-stimulated genes, such as IFN-β/λ, ISG15, CXCL11, CXCL3 and IL-6, etc. Moreover, infected hBMECs exhibited a disruption in the cytoskeletal structure both on the transcriptomic and cytological levels. The RNAseq analysis showed different pathways and candidate genes associated with the neuroactive ligand-receptor interaction, neuroinflammation, and neurodegenerative diseases, together with a predicted activation of the neuroglia. Likewise, some genes linked with the mitochondrial structure and function displayed a significantly altered expression. En masse, this data supports that hBMECs could be infected by the IAV, which induces the innate and inflammatory immune response. The results suggest that the influenza virus infection could potentially induce a subsequent aggravation of neurological disorders.
Supplementary information: The online version contains supplementary material available at 10.1186/s44149-022-00053-9.
Keywords: Blood–brain barrier; CNS; Influenza A virus (IAV); Neurodegenerative diseases; RNAseq; hBMECs.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsAuthor Min Cui was not involved in the journal’s review or decisions related to this manuscript. The authors declare that they have no competing interests.
Figures







Similar articles
-
Zika Virus Persistently Infects and Is Basolaterally Released from Primary Human Brain Microvascular Endothelial Cells.mBio. 2017 Jul 11;8(4):e00952-17. doi: 10.1128/mBio.00952-17. mBio. 2017. PMID: 28698279 Free PMC article.
-
Innate Immune Response to Influenza Virus at Single-Cell Resolution in Human Epithelial Cells Revealed Paracrine Induction of Interferon Lambda 1.J Virol. 2019 Sep 30;93(20):e00559-19. doi: 10.1128/JVI.00559-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31375585 Free PMC article.
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Immune response and blood-brain barrier dysfunction during viral neuroinvasion.Innate Immun. 2021 Feb;27(2):109-117. doi: 10.1177/1753425920954281. Epub 2020 Sep 9. Innate Immun. 2021. PMID: 32903111 Free PMC article. Review.
Cited by
-
Risk factors for death associated with severe influenza in children and the impact of the COVID-19 pandemic on clinical characteristics.Front Pediatr. 2023 Sep 12;11:1249058. doi: 10.3389/fped.2023.1249058. eCollection 2023. Front Pediatr. 2023. PMID: 37772040 Free PMC article.
-
Innate immune sensors and regulators at the blood brain barrier: focus on toll-like receptors and inflammasomes as mediators of neuro-immune crosstalk and inflammation.J Neuroinflammation. 2025 Feb 15;22(1):39. doi: 10.1186/s12974-025-03360-3. J Neuroinflammation. 2025. PMID: 39955600 Free PMC article. Review.
-
Type I/type III IFN and related factors regulate JEV infection and BBB endothelial integrity.J Neuroinflammation. 2023 Sep 27;20(1):216. doi: 10.1186/s12974-023-02891-x. J Neuroinflammation. 2023. PMID: 37752509 Free PMC article.
-
Interferon-induced protein ISG15 in the central nervous system, quo vadis?FEBS Lett. 2025 May 12:10.1002/1873-3468.70063. doi: 10.1002/1873-3468.70063. Online ahead of print. FEBS Lett. 2025. PMID: 40353372 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
