Distinct pathogenic roles for resident and monocyte-derived macrophages in lupus nephritis
- PMID: 36345939
- PMCID: PMC9675473
- DOI: 10.1172/jci.insight.159751
Distinct pathogenic roles for resident and monocyte-derived macrophages in lupus nephritis
Abstract
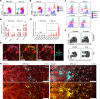
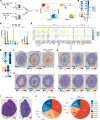
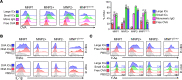
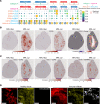
Lupus nephritis is a serious complication of systemic lupus erythematosus, mediated by IgG immune complex (IC) deposition in kidneys, with limited treatment options. Kidney macrophages are critical tissue sentinels that express IgG-binding Fcγ receptors (FcγRs), with previous studies identifying prenatally seeded resident macrophages as major IC responders. Using single-cell transcriptomic and spatial analyses in murine and human lupus nephritis, we sought to understand macrophage heterogeneity and subset-specific contributions in disease. In lupus nephritis, the cell fate trajectories of tissue-resident (TrMac) and monocyte-derived (MoMac) kidney macrophages were perturbed, with disease-associated transcriptional states indicating distinct pathogenic roles for TrMac and MoMac subsets. Lupus nephritis-associated MoMac subsets showed marked induction of FcγR response genes, avidly internalized circulating ICs, and presented IC-opsonized antigen. In contrast, lupus nephritis-associated TrMac subsets demonstrated limited IC uptake, but expressed monocyte chemoattractants, and their depletion attenuated monocyte recruitment to the kidney. TrMacs also produced B cell tissue niche factors, suggesting a role in supporting autoantibody-producing lymphoid aggregates. Extensive similarities were observed with human kidney macrophages, revealing cross-species transcriptional disruption in lupus nephritis. Overall, our study suggests a division of labor in the kidney macrophage response in lupus nephritis, with treatment implications - TrMacs orchestrate leukocyte recruitment while MoMacs take up and present IC antigen.
Keywords: Autoimmunity; Immunology; Lupus; Macrophages.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

