Two neuronal peptides encoded from a single transcript regulate mitochondrial complex III in Drosophila
- PMID: 36346220
- PMCID: PMC9681215
- DOI: 10.7554/eLife.82709
Two neuronal peptides encoded from a single transcript regulate mitochondrial complex III in Drosophila
Abstract
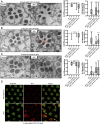
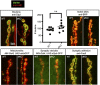
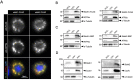
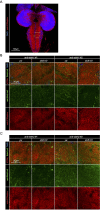
Naturally produced peptides (<100 amino acids) are important regulators of physiology, development, and metabolism. Recent studies have predicted that thousands of peptides may be translated from transcripts containing small open-reading frames (smORFs). Here, we describe two peptides in Drosophila encoded by conserved smORFs, Sloth1 and Sloth2. These peptides are translated from the same bicistronic transcript and share sequence similarities, suggesting that they encode paralogs. Yet, Sloth1 and Sloth2 are not functionally redundant, and loss of either peptide causes animal lethality, reduced neuronal function, impaired mitochondrial function, and neurodegeneration. We provide evidence that Sloth1/2 are highly expressed in neurons, imported to mitochondria, and regulate mitochondrial complex III assembly. These results suggest that phenotypic analysis of smORF genes in Drosophila can provide a wealth of information on the biological functions of this poorly characterized class of genes.
Keywords: CRISPR/Cas9; D. melanogaster; genetics; genomics; mitochondria; neurodegeneration; neuroscience; paralogs; peptide; smORF.
Conflict of interest statement
JB, BU, IP, JR, FE, ZZ, BB, SC, DS, NP No competing interests declared, HB Reviewing editor, eLife
Figures















Similar articles
-
Molecular and functional characterization of the Drosophila melanogaster conserved smORFome.Cell Rep. 2023 Nov 28;42(11):113311. doi: 10.1016/j.celrep.2023.113311. Epub 2023 Oct 26. Cell Rep. 2023. PMID: 37889754 Free PMC article.
-
Small ORFs, Big Insights: Drosophila as a Model to Unraveling Microprotein Functions.Cells. 2024 Oct 2;13(19):1645. doi: 10.3390/cells13191645. Cells. 2024. PMID: 39404408 Free PMC article. Review.
-
Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq.Elife. 2014 Aug 21;3:e03528. doi: 10.7554/eLife.03528. Elife. 2014. PMID: 25144939 Free PMC article.
-
Classification and function of small open reading frames.Nat Rev Mol Cell Biol. 2017 Sep;18(9):575-589. doi: 10.1038/nrm.2017.58. Epub 2017 Jul 12. Nat Rev Mol Cell Biol. 2017. PMID: 28698598
-
Small open reading frames and cellular stress responses.Mol Omics. 2019 Apr 1;15(2):108-116. doi: 10.1039/c8mo00283e. Epub 2019 Feb 27. Mol Omics. 2019. PMID: 30810554 Free PMC article. Review.
Cited by
-
Molecular and functional characterization of the Drosophila melanogaster conserved smORFome.Cell Rep. 2023 Nov 28;42(11):113311. doi: 10.1016/j.celrep.2023.113311. Epub 2023 Oct 26. Cell Rep. 2023. PMID: 37889754 Free PMC article.
-
An intermembrane space protein facilitates completion of mitochondrial division in yeast.bioRxiv [Preprint]. 2023 Mar 31:2023.03.31.535139. doi: 10.1101/2023.03.31.535139. bioRxiv. 2023. Update in: J Cell Biol. 2023 Oct 2;222(10):e202303147. doi: 10.1083/jcb.202303147. PMID: 37034761 Free PMC article. Updated. Preprint.
-
Mitochondrial Neurodegeneration: Lessons from Drosophila melanogaster Models.Biomolecules. 2023 Feb 16;13(2):378. doi: 10.3390/biom13020378. Biomolecules. 2023. PMID: 36830747 Free PMC article. Review.
-
Small ORFs, Big Insights: Drosophila as a Model to Unraveling Microprotein Functions.Cells. 2024 Oct 2;13(19):1645. doi: 10.3390/cells13191645. Cells. 2024. PMID: 39404408 Free PMC article. Review.
-
The pleiotropic functions of Pri smORF peptides synchronize leg development regulators.PLoS Genet. 2023 Oct 30;19(10):e1011004. doi: 10.1371/journal.pgen.1011004. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37903161 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

