Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-Mutant Pancreatic Cancer
- PMID: 36346366
- PMCID: PMC9812941
- DOI: 10.1158/0008-5472.CAN-22-0391
Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-Mutant Pancreatic Cancer
Abstract
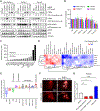
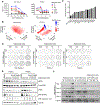
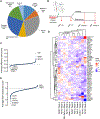
Mutational loss of CDKN2A (encoding p16INK4A) tumor-suppressor function is a key genetic step that complements activation of KRAS in promoting the development and malignant growth of pancreatic ductal adenocarcinoma (PDAC). However, pharmacologic restoration of p16INK4A function with inhibitors of CDK4 and CDK6 (CDK4/6) has shown limited clinical efficacy in PDAC. Here, we found that concurrent treatment with both a CDK4/6 inhibitor (CDK4/6i) and an ERK-MAPK inhibitor (ERKi) synergistically suppresses the growth of PDAC cell lines and organoids by cooperatively blocking CDK4/6i-induced compensatory upregulation of ERK, PI3K, antiapoptotic signaling, and MYC expression. On the basis of these findings, a Phase I clinical trial was initiated to evaluate the ERKi ulixertinib in combination with the CDK4/6i palbociclib in patients with advanced PDAC (NCT03454035). As inhibition of other proteins might also counter CDK4/6i-mediated signaling changes to increase cellular CDK4/6i sensitivity, a CRISPR-Cas9 loss-of-function screen was conducted that revealed a spectrum of functionally diverse genes whose loss enhanced CDK4/6i growth inhibitory activity. These genes were enriched around diverse signaling nodes, including cell-cycle regulatory proteins centered on CDK2 activation, PI3K-AKT-mTOR signaling, SRC family kinases, HDAC proteins, autophagy-activating pathways, chromosome regulation and maintenance, and DNA damage and repair pathways. Novel therapeutic combinations were validated using siRNA and small-molecule inhibitor-based approaches. In addition, genes whose loss imparts a survival advantage were identified (e.g., RB1, PTEN, FBXW7), suggesting possible resistance mechanisms to CDK4/6 inhibition. In summary, this study has identified novel combinations with CDK4/6i that may have clinical benefit to patients with PDAC.
Significance: CRISPR-Cas9 screening and protein activity mapping reveal combinations that increase potency of CDK4/6 inhibitors and overcome drug-induced compensations in pancreatic cancer.
©2022 American Association for Cancer Research.
Conflict of interest statement
Authors’ Disclosures
C.J. Der is a consultant/advisory board member for Anchiano Therapeutics, Deciphera Pharmaceuticals, Mirati Therapeutics and Revolution Medicines. C.J. Der has received research funding support from SpringWorks Therapeutics, Mirati Therapeutics and Deciphera Pharmaceuticals, and has consulted for Eli Lilly, Jazz Therapeutics, Ribometrix, Sanofi, and Turning Point Therapeutics. A.D. Cox has consulted for Eli Lilly and Mirati Therapeutics. K.L. Bryant has received research funding support from SpringWorks Therapeutics. M. Pierobon and E.F. Petricoin are inventors on US Government and University assigned patents and patent applications that cover aspects of the technologies discussed such as the Reverse Phase Protein Microarrays. As inventors, they are entitled to receive royalties as provided by US Law and George Mason University policy. M. Pierobon and E.F. Petricoin receive royalties from and are consultants of TheraLink Technologies, Inc. E.F. Petricoin is a shareholder of TheraLink Technologies, Inc and a shareholder and consultant of Perthera, Inc. J. Sorrentino was an employee of G1 Therapeutics while the work in this manuscript was completed. A. Beelen is an employee of G1 Therapeutics. All other authors declare no potential conflicts of interest.
Figures




References
-
- Hayashi A, Hong J, Iacobuzio-Donahue CA. The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol 2021;18:469–81 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

