NBN Pathogenic Germline Variants are Associated with Pan-Cancer Susceptibility and In Vitro DNA Damage Response Defects
- PMID: 36346689
- PMCID: PMC9843434
- DOI: 10.1158/1078-0432.CCR-22-1703
NBN Pathogenic Germline Variants are Associated with Pan-Cancer Susceptibility and In Vitro DNA Damage Response Defects
Abstract
Purpose: To explore the role of NBN as a pan-cancer susceptibility gene.
Experimental design: Matched germline and somatic DNA samples from 34,046 patients were sequenced using Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets and presumed pathogenic germline variants (PGV) identified. Allele-specific and gene-centered analysis of enrichment was conducted and a validation cohort of 26,407 pan-cancer patients was analyzed. Functional studies utilized cellular models with analysis of protein expression, MRN complex formation/localization, and viability assessment following treatment with γ-irradiation.
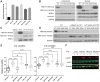
Results: We identified 83 carriers of 32 NBN PGVs (0.25% of the studied series), 40% of which (33/83) carried the Slavic founder p.K219fs. The frequency of PGVs varied across cancer types. Patients harboring NBN PGVs demonstrated increased loss of the wild-type allele in their tumors [OR = 2.7; confidence interval (CI): 1.4-5.5; P = 0.0024; pan-cancer], including lung and pancreatic tumors compared with breast and colorectal cancers. p.K219fs was enriched across all tumor types (OR = 2.22; CI: 1.3-3.6; P = 0.0018). Gene-centered analysis revealed enrichment of PGVs in cases compared with controls in the European population (OR = 1.9; CI: 1.3-2.7; P = 0.0004), a finding confirmed in the replication cohort (OR = 1.8; CI: 1.2-2.6; P = 0.003). Two novel truncating variants, p.L19* and p.N71fs, produced a 45 kDa fragment generated by alternative translation initiation that maintained binding to MRE11. Cells expressing these fragments showed higher sensitivity to γ-irradiation and lower levels of radiation-induced KAP1 phosphorylation.
Conclusions: Burden analyses, biallelic inactivation, and functional evidence support the role of NBN as contributing to a broad cancer spectrum. Further studies in large pan-cancer series and the assessment of epistatic and environmental interactions are warranted to further define these associations.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Weemaes CM, Hustinx TW, Scheres JM, van Munster PJ, Bakkeren JA, Taalman RD. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand 1981;70:557–64. - PubMed
-
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, et al. . The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 1998;93:477–86. - PubMed
-
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, et al. . Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 1998;93:467–76. - PubMed
-
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair 2004;3:1207–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

