Hippo signaling pathway activation during SARS-CoV-2 infection contributes to host antiviral response
- PMID: 36346780
- PMCID: PMC9642871
- DOI: 10.1371/journal.pbio.3001851
Hippo signaling pathway activation during SARS-CoV-2 infection contributes to host antiviral response
Abstract
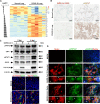
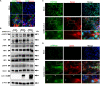
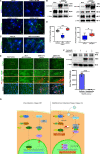
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), responsible for the Coronavirus Disease 2019 (COVID-19) pandemic, causes respiratory failure and damage to multiple organ systems. The emergence of viral variants poses a risk of vaccine failures and prolongation of the pandemic. However, our understanding of the molecular basis of SARS-CoV-2 infection and subsequent COVID-19 pathophysiology is limited. In this study, we have uncovered a critical role for the evolutionarily conserved Hippo signaling pathway in COVID-19 pathogenesis. Given the complexity of COVID-19-associated cell injury and immunopathogenesis processes, we investigated Hippo pathway dynamics in SARS-CoV-2 infection by utilizing COVID-19 lung samples and human cell models based on pluripotent stem cell-derived cardiomyocytes (PSC-CMs) and human primary lung air-liquid interface (ALI) cultures. SARS-CoV-2 infection caused activation of the Hippo signaling pathway in COVID-19 lung and in vitro cultures. Both parental and Delta variant of concern (VOC) strains induced Hippo pathway. The chemical inhibition and gene knockdown of upstream kinases MST1/2 and LATS1 resulted in significantly enhanced SARS-CoV-2 replication, indicating antiviral roles. Verteporfin, a pharmacological inhibitor of the Hippo pathway downstream transactivator, YAP, significantly reduced virus replication. These results delineate a direct antiviral role for Hippo signaling in SARS-CoV-2 infection and the potential for this pathway to be pharmacologically targeted to treat COVID-19.
Copyright: © 2022 Garcia et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Update of
-
Hippo Signaling Pathway Activation during SARS-CoV-2 Infection Contributes to Host Antiviral Response.bioRxiv [Preprint]. 2022 Apr 8:2022.04.07.487520. doi: 10.1101/2022.04.07.487520. bioRxiv. 2022. Update in: PLoS Biol. 2022 Nov 8;20(11):e3001851. doi: 10.1371/journal.pbio.3001851. PMID: 35441167 Free PMC article. Updated. Preprint.
References
-
- Ramaiah A, Arumugaswami V. Insights into Cross-species Evolution of Novel Human Coronavirus 2019-nCoV and Defining Immune Determinants for Vaccine Development. bioRxiv. 2020:2020.01.29.925867.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

