Distinct Genetically Determined Origins of Myd88/BCL2-Driven Aggressive Lymphoma Rationalize Targeted Therapeutic Intervention Strategies
- PMID: 36346827
- PMCID: PMC9816818
- DOI: 10.1158/2643-3230.BCD-22-0007
Distinct Genetically Determined Origins of Myd88/BCL2-Driven Aggressive Lymphoma Rationalize Targeted Therapeutic Intervention Strategies
Abstract
Genomic profiling revealed the identity of at least 5 subtypes of diffuse large B-cell lymphoma (DLBCL), including the MCD/C5 cluster characterized by aberrations in MYD88, BCL2, PRDM1, and/or SPIB. We generated mouse models harboring B cell-specific Prdm1 or Spib aberrations on the background of oncogenic Myd88 and Bcl2 lesions. We deployed whole-exome sequencing, transcriptome, flow-cytometry, and mass cytometry analyses to demonstrate that Prdm1- or Spib-altered lymphomas display molecular features consistent with prememory B cells and light-zone B cells, whereas lymphomas lacking these alterations were enriched for late light-zone and plasmablast-associated gene sets. Consistent with the phenotypic evidence for increased B cell receptor signaling activity in Prdm1-altered lymphomas, we demonstrate that combined BTK/BCL2 inhibition displays therapeutic activity in mice and in five of six relapsed/refractory DLBCL patients. Moreover, Prdm1-altered lymphomas were immunogenic upon transplantation into immuno-competent hosts, displayed an actionable PD-L1 surface expression, and were sensitive to antimurine-CD19-CAR-T cell therapy, in vivo.
Significance: Relapsed/refractory DLBCL remains a major medical challenge, and most of these patients succumb to their disease. Here, we generated mouse models, faithfully recapitulating the biology of MYD88-driven human DLBCL. These models revealed robust preclinical activity of combined BTK/BCL2 inhibition. We confirmed activity of this regimen in pretreated non-GCB-DLBCL patients. See related commentary by Leveille et al., p. 8. This article is highlighted in the In This Issue feature, p. 1.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures
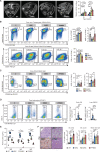


![Figure 4. Murine PPMBC lymphomas are susceptible to immune-therapeutic approaches. A, Schematic depiction of the anti-mCD19scFv-CD28-CD3z CAR construct (27). B, Population sizes of mock- and CAR-transduced T cells isolated from C57BL6/J wild-type spleens and a CD19+ PPMBC lymphoma cell line at the indicated effector:target ratios after 48 hours of coculture after gating for live cells. C, PPMBC mice with a low or high lymphoma-burden [spleen volume 500–700 μL (n = 3) or >700 μL (n = 3), respectively] were treated with a single injection of 2 × 106 anti-mCD19 CAR-T cells. Tumor volume was monitored weekly via MRI and is visualized as fold change from baseline. D, Kaplan–Meier curve showing the overall survival of CAR-T–treated mice and untreated controls. The CAR-T–treated cohort is depicted in total, as well as separated into high- and low lymphoma-burden animals. E, MRI-determined tumor volume of PPMBC animals treated with an anti–PD-L1 (n = 7), displayed as fold change from baseline. Untreated animals (n = 8) served as controls. F, Kaplan–Meier curve showing the overall survival of anti–PD-L1-treated mice (n = 7) and untreated controls (n = 8). *, P ≤ 0.05; **, P ≤ 0.01; log-rank test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/7d2d/9816818/41d6718070e3/78fig4.gif)

Comment in
-
Genetic Modeling of B-cell State Transitions for Rational Design of Lymphoma Therapies.Blood Cancer Discov. 2023 Jan 6;4(1):8-11. doi: 10.1158/2643-3230.BCD-22-0180. Blood Cancer Discov. 2023. PMID: 36534735 Free PMC article.
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. - PubMed
-
- Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol 2015;15:172–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

