RASopathy mutations provide functional insight into the BRAF cysteine-rich domain and reveal the importance of autoinhibition in BRAF regulation
- PMID: 36347258
- PMCID: PMC9677513
- DOI: 10.1016/j.molcel.2022.10.016
RASopathy mutations provide functional insight into the BRAF cysteine-rich domain and reveal the importance of autoinhibition in BRAF regulation
Abstract
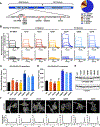
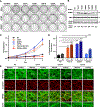
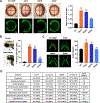
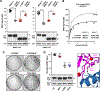
BRAF is frequently mutated in human cancer and the RASopathy syndromes, with RASopathy mutations often observed in the cysteine-rich domain (CRD). Although the CRD participates in phosphatidylserine (PS) binding, the RAS-RAF interaction, and RAF autoinhibition, the impact of these activities on RAF function in normal and disease states is not well characterized. Here, we analyze a panel of CRD mutations and show that they increase BRAF activity by relieving autoinhibition and/or enhancing PS binding, with relief of autoinhibition being the major factor determining mutation severity. Further, we show that CRD-mediated autoinhibition prevents the constitutive plasma membrane localization of BRAF that causes increased RAS-dependent and RAS-independent function. Comparison of the BRAF- and CRAF-CRDs also indicates that the BRAF-CRD is a stronger mediator of autoinhibition and PS binding, and given the increased catalytic activity of BRAF, our studies reveal a more critical role for CRD-mediated autoinhibition in BRAF regulation.
Keywords: BRAF; CRAF; CRD; RAF kinases; RAS; RASopathies; autoinhibition; cysteine-rich domain; development; phosphatidylserine.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
RASopathy mutations open new insights into the mechanism of BRAF activation.Mol Cell. 2022 Nov 17;82(22):4192-4193. doi: 10.1016/j.molcel.2022.10.034. Mol Cell. 2022. PMID: 36400004 Free PMC article.
References
-
- Chong H, and Guan K-L (2003). Regulation of Raf through phosphorylation and N-terminus-C terminus interaction. J. Biol. Chem. 278, 36269–36276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

