T cell responses at diagnosis of amyotrophic lateral sclerosis predict disease progression
- PMID: 36347843
- PMCID: PMC9643478
- DOI: 10.1038/s41467-022-34526-9
T cell responses at diagnosis of amyotrophic lateral sclerosis predict disease progression
Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease, involving neuroinflammation and T cell infiltration in the central nervous system. However, the contribution of T cell responses to the pathology of the disease is not fully understood. Here we show, by flow cytometric analysis of blood and cerebrospinal fluid (CSF) samples of a cohort of 89 newly diagnosed ALS patients in Stockholm, Sweden, that T cell phenotypes at the time of diagnosis are good predictors of disease outcome. High frequency of CD4+FOXP3- effector T cells in blood and CSF is associated with poor survival, whereas high frequency of activated regulatory T (Treg) cells and high ratio between activated and resting Treg cells in blood are associated with better survival. Besides survival, phenotypic profiling of T cells could also predict disease progression rate. Single cell transcriptomics analysis of CSF samples shows clonally expanded CD4+ and CD8+ T cells in CSF, with characteristic gene expression patterns. In summary, T cell responses associate with and likely contribute to disease progression in ALS, supporting modulation of adaptive immunity as a viable therapeutic option.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
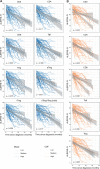

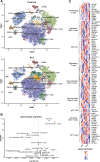


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

