GZMKhigh CD8+ T effector memory cells are associated with CD15high neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome
- PMID: 36347862
- PMCID: PMC9643357
- DOI: 10.1038/s41467-022-34467-3
GZMKhigh CD8+ T effector memory cells are associated with CD15high neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome
Erratum in
-
Author Correction: GZMKhigh CD8+ T effector memory cells are associated with CD15high neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome.Nat Commun. 2022 Nov 30;13(1):7380. doi: 10.1038/s41467-022-35251-z. Nat Commun. 2022. PMID: 36450724 Free PMC article. No abstract available.
Abstract
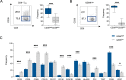
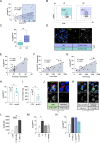
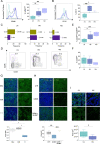
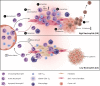
CD8+ T cells are a major prognostic determinant in solid tumors, including colorectal cancer (CRC). However, understanding how the interplay between different immune cells impacts on clinical outcome is still in its infancy. Here, we describe that the interaction of tumor infiltrating neutrophils expressing high levels of CD15 with CD8+ T effector memory cells (TEM) correlates with tumor progression. Mechanistically, stromal cell-derived factor-1 (CXCL12/SDF-1) promotes the retention of neutrophils within tumors, increasing the crosstalk with CD8+ T cells. As a consequence of the contact-mediated interaction with neutrophils, CD8+ T cells are skewed to produce high levels of GZMK, which in turn decreases E-cadherin on the intestinal epithelium and favors tumor progression. Overall, our results highlight the emergence of GZMKhigh CD8+ TEM in non-metastatic CRC tumors as a hallmark driven by the interaction with neutrophils, which could implement current patient stratification and be targeted by novel therapeutics.
© 2022. The Author(s).
Conflict of interest statement
E.L. receives research grants from Brystol Myers Squibb on a topic unrelated to this work and serves as a consultant for BD Biosciences. The remaining authors declare no competing interests.
Figures



Similar articles
-
Author Correction: GZMKhigh CD8+ T effector memory cells are associated with CD15high neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome.Nat Commun. 2022 Nov 30;13(1):7380. doi: 10.1038/s41467-022-35251-z. Nat Commun. 2022. PMID: 36450724 Free PMC article. No abstract available.
-
The Interplay Between Neutrophils and CD8+ T Cells Improves Survival in Human Colorectal Cancer.Clin Cancer Res. 2017 Jul 15;23(14):3847-3858. doi: 10.1158/1078-0432.CCR-16-2047. Epub 2017 Jan 20. Clin Cancer Res. 2017. PMID: 28108544
-
Intratumoral infiltrating lymphocytes correlate with improved survival in colorectal cancer patients: Independent of oncogenetic features.J Cell Physiol. 2019 Apr;234(4):4768-4777. doi: 10.1002/jcp.27273. Epub 2018 Oct 28. J Cell Physiol. 2019. PMID: 30370522
-
The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin.Front Immunol. 2018 Aug 15;9:1904. doi: 10.3389/fimmu.2018.01904. eCollection 2018. Front Immunol. 2018. PMID: 30158938 Free PMC article. Review.
-
Immune scores in colorectal cancer: Where are we?Eur J Cancer. 2020 Nov;140:105-118. doi: 10.1016/j.ejca.2020.08.024. Epub 2020 Oct 16. Eur J Cancer. 2020. PMID: 33075623 Review.
Cited by
-
Transcriptomic analysis reveals the potential crosstalk genes and immune relationship between IgA nephropathy and periodontitis.Front Immunol. 2023 Jan 30;14:1062590. doi: 10.3389/fimmu.2023.1062590. eCollection 2023. Front Immunol. 2023. PMID: 36793719 Free PMC article.
-
Maintenance of caecal homeostasis by diverse adaptive immune cells in the rhesus macaque.Clin Transl Immunology. 2024 May 2;13(5):e1508. doi: 10.1002/cti2.1508. eCollection 2024. Clin Transl Immunology. 2024. PMID: 38707998 Free PMC article.
-
scRNA-seq reveals an immune microenvironment and JUN-mediated NK cell exhaustion in relapsed T-ALL.Cell Rep Med. 2025 May 20;6(5):102098. doi: 10.1016/j.xcrm.2025.102098. Epub 2025 Apr 29. Cell Rep Med. 2025. PMID: 40306275 Free PMC article.
-
Age-Associated Modulation of TREM1/2-Expressing Macrophages Promotes Melanoma Progression and Metastasis.Cancer Res. 2025 Jun 16;85(12):2218-2233. doi: 10.1158/0008-5472.CAN-24-4317. Cancer Res. 2025. PMID: 40126543 Free PMC article.
-
A novel stratification framework based on anoikis-related genes for predicting the prognosis in patients with osteosarcoma.Front Immunol. 2023 Jul 27;14:1199869. doi: 10.3389/fimmu.2023.1199869. eCollection 2023. Front Immunol. 2023. PMID: 37575253 Free PMC article.
References
-
- American Cancer Society. Colorectal Cancer Fact and Figures. www.cancer.org (2020–2022)
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

