The first mitotic division of human embryos is highly error prone
- PMID: 36347869
- PMCID: PMC9643329
- DOI: 10.1038/s41467-022-34294-6
The first mitotic division of human embryos is highly error prone
Abstract
Human beings are made of ~50 trillion cells which arise from serial mitotic divisions of a single cell - the fertilised egg. Remarkably, the early human embryo is often chromosomally abnormal, and many are mosaic, with the karyotype differing from one cell to another. Mosaicism presumably arises from chromosome segregation errors during the early mitotic divisions, although these events have never been visualised in living human embryos. Here, we establish live cell imaging of chromosome segregation using normally fertilised embryos from an egg-share-to-research programme, as well as embryos deselected during fertility treatment. We reveal that the first mitotic division has an extended prometaphase/metaphase and exhibits phenotypes that can cause nondisjunction. These included multipolar chromosome segregations and lagging chromosomes that lead to formation of micronuclei. Analysis of nuclear number and size provides evidence of equivalent phenotypes in 2-cell human embryos that gave rise to live births. Together this shows that errors in the first mitotic division can be tolerated in human embryos and uncovers cell biological events that contribute to preimplantation mosaicism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
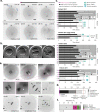
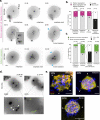


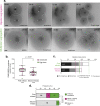
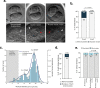
References
-
- Boklage CE. Survival probability of human conceptions from fertilization to term. Int. J. Fertil. 1990;35:75–94. - PubMed
-
- Soler A, et al. Overview of chromosome abnormalities in first trimester miscarriages: a series of 1,011 consecutive chorionic villi sample karyotypes. Cytogenet. Genome Res. 2017;152:81–89. - PubMed
-
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

