Deep learning algorithm reveals probabilities of stage-specific time to conversion in individuals with neurodegenerative disease LATE
- PMID: 36348767
- PMCID: PMC9632667
- DOI: 10.1002/trc2.12363
Deep learning algorithm reveals probabilities of stage-specific time to conversion in individuals with neurodegenerative disease LATE
Abstract
Introduction: Limbic-predominant age-related TAR DNA-binding protein 43 (TDP-43) encephalopathy (LATE) is a recently defined neurodegenerative disease. Currently, there is no effective way to make a prognosis of time to stage-specific future conversions at an individual level.
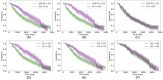
Methods: After using the Kaplan-Meier estimation and log-rank test to confirm the heterogeneity of LATE progression, we developed a deep learning-based approach to assess the stage-specific probabilities of time to LATE conversions for different subjects.
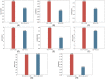
Results: Our approach could accurately estimate the disease incidence and transition to next stages: the concordance index was at least 82% and the integrated Brier score was less than 0.14. Moreover, we identified the top 10 important predictors for each disease conversion scenario to help explain the estimation results, which were clinicopathologically meaningful and most were also statistically significant.
Discussion: Our study has the potential to provide individualized assessment for future time courses of LATE conversions years before their actual occurrence.
Keywords: limbic‐predominant age‐related TAR DNA‐binding protein 43 encephalopathy; machine learning; progression rate; stage‐stratified analysis; survival models; time‐to‐event estimation.
© 2022 The Authors. Alzheimer's & Dementia: Translational Research & Clinical Interventions published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
The authors declare no conflicts of interest. Author disclosures are available in the supporting information.
Figures


References
Grants and funding
- P30 AG072946/AG/NIA NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- P50 AG047266/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R21 AG070909/AG/NIA NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- P50 AG047366/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- R01 HD101508/HD/NICHD NIH HHS/United States
- P30 AG028383/AG/NIA NIH HHS/United States
- P50 AG033514/AG/NIA NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- P30 AG053760/AG/NIA NIH HHS/United States
- R56 NS117587/NS/NINDS NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- P50 AG023501/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- P30 AG035982/AG/NIA NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- P30 AG008051/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- P50 AG047270/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P30 AG049638/AG/NIA NIH HHS/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- P30 AG008017/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
