Sparse prediction informed by genetic annotations using the logit normal prior for Bayesian regression tree ensembles
- PMID: 36349692
- PMCID: PMC9892284
- DOI: 10.1002/gepi.22505
Sparse prediction informed by genetic annotations using the logit normal prior for Bayesian regression tree ensembles
Abstract
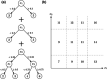
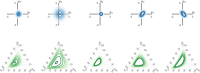
Using high-dimensional genetic variants such as single nucleotide polymorphisms (SNP) to predict complex diseases and traits has important applications in basic research and other clinical settings. For example, predicting gene expression is a necessary first step to identify (putative) causal genes in transcriptome-wide association studies. Due to weak signals, high-dimensionality, and linkage disequilibrium (correlation) among SNPs, building such a prediction model is challenging. However, functional annotations at the SNP level (e.g., as epigenomic data across multiple cell- or tissue-types) are available and could be used to inform predictor importance and aid in outcome prediction. Existing approaches to incorporate annotations have been based mainly on (generalized) linear models. Bayesian additive regression trees (BART), in contrast, is a reliable method to obtain high-quality nonlinear out of sample predictions without overfitting. Unfortunately, the default prior from BART may be too inflexible to handle sparse situations where the number of predictors approaches or surpasses the number of observations. Motivated by our real data application, this article proposes an alternative prior based on the logit normal distribution because it provides a framework that is adaptive to sparsity and can model informative functional annotations. It also provides a framework to incorporate prior information about the between SNP correlations. Computational details for carrying out inference are presented along with the results from a simulation study and a genome-wide prediction analysis of the Alzheimer's Disease Neuroimaging Initiative data.
Keywords: ensemble learning; genetics; high-dimensional prediction; sparsity.
© 2022 The Authors. Genetic Epidemiology published by Wiley Periodicals LLC.
Figures


References
-
- Albert, J. , & Chib, S. (1993). Bayesian analysis of binary and polychotomous response data. Journal of the American Statistical Association, 88(422), 669–679. 10.1080/01621459.1993.10476321 - DOI
-
- Bishop, C. M. (2006). Pattern recognition and machine learning. Springer‐Verlag.
-
- Bleich, J. , Kapelner, A. , George, E. , & Jensen, S. (2014). Variable selection for BART: An application to gene regulation. The Annals of Applied Statistics, 8(3), 1750–1781. 10.1214/14-aoas755 - DOI
-
- Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32. 10.1023/a:1010933404324 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

