A Phase I/II Open-Label Study of Molibresib for the Treatment of Relapsed/Refractory Hematologic Malignancies
- PMID: 36350312
- PMCID: PMC9932578
- DOI: 10.1158/1078-0432.CCR-22-1284
A Phase I/II Open-Label Study of Molibresib for the Treatment of Relapsed/Refractory Hematologic Malignancies
Abstract
Purpose: Molibresib is a selective, small molecule inhibitor of the bromodomain and extra-terminal (BET) protein family. This was an open-label, two-part, Phase I/II study investigating molibresib monotherapy for the treatment of hematological malignancies (NCT01943851).
Patients and methods: Part 1 (dose escalation) determined the recommended Phase 2 dose (RP2D) of molibresib in patients with acute myeloid leukemia (AML), Non-Hodgkin lymphoma (NHL), or multiple myeloma. Part 2 (dose expansion) investigated the safety and efficacy of molibresib at the RP2D in patients with relapsed/refractory myelodysplastic syndrome (MDS; as well as AML evolved from antecedent MDS) or cutaneous T-cell lymphoma (CTCL). The primary endpoint in Part 1 was safety and the primary endpoint in Part 2 was objective response rate (ORR).
Results: There were 111 patients enrolled (87 in Part 1, 24 in Part 2). Molibresib RP2Ds of 75 mg daily (for MDS) and 60 mg daily (for CTCL) were selected. Most common Grade 3+ adverse events included thrombocytopenia (37%), anemia (15%), and febrile neutropenia (15%). Six patients achieved complete responses [3 in Part 1 (2 AML, 1 NHL), 3 in Part 2 (MDS)], and 7 patients achieved partial responses [6 in Part 1 (4 AML, 2 NHL), 1 in Part 2 (MDS)]. The ORRs for Part 1, Part 2, and the total study population were 10% [95% confidence interval (CI), 4.8-18.7], 25% (95% CI, 7.3-52.4), and 13% (95% CI, 6.9-20.6), respectively.
Conclusions: While antitumor activity was observed with molibresib, use was limited by gastrointestinal and thrombocytopenia toxicities. Investigations of molibresib as part of combination regimens may be warranted.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures
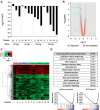
![Figures 3. A and B, Heatmaps of individual patients’ change in gene expression following molibresib treatment (log2; fold change) for genes that were identified as differentially expressed when pooled across all NRs (A), or a clinical response (CR and PR; B). Genes are defined as differentially expressed if they reach a 1.5-fold change with an FDR <0.05. The bar at the top of the heatmap corresponds to clinical response, indicating CR [pink; Patient 1 with CRp, Patient 2 with CRi), PR (blue), or NR (green)]. C, Venn diagram showing the number of differentially expressed genes in responders, non-responder, or both. D, Scatterplot showing the fold change in gene expression for the two patients that achieved CR. Genes are categorized as unchanged in both patients (gray), differentially expressed in one patient (blue), differentially expressed in the same direction in both patients (black), or differentially expressed in both patients, but in the opposite direction (red) if they exhibited >1.5-fold change (dotted lines). CR, complete response; CRi, CR but platelet count <100×109/L or neutrophil count <1×109/L; CRp, CR but platelet count <100×109/L; FDR, false discovery rate; NRs, non-responders; PR, partial response.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/f715/9932578/30188bce2e6d/711fig3.gif)
Comment in
- 1078-0432. doi: 10.1158/1078-0432.CCR-29-4-HI doi: 10.1158/1078-0432.CCR-29-4-HI
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

