The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource
- PMID: 36350656
- PMCID: PMC9825413
- DOI: 10.1093/nar/gkac1010
The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource
Abstract
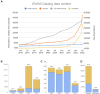
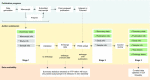
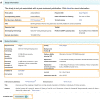
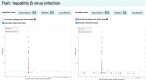
The NHGRI-EBI GWAS Catalog (www.ebi.ac.uk/gwas) is a FAIR knowledgebase providing detailed, structured, standardised and interoperable genome-wide association study (GWAS) data to >200 000 users per year from academic research, healthcare and industry. The Catalog contains variant-trait associations and supporting metadata for >45 000 published GWAS across >5000 human traits, and >40 000 full P-value summary statistics datasets. Content is curated from publications or acquired via author submission of prepublication summary statistics through a new submission portal and validation tool. GWAS data volume has vastly increased in recent years. We have updated our software to meet this scaling challenge and to enable rapid release of submitted summary statistics. The scope of the repository has expanded to include additional data types of high interest to the community, including sequencing-based GWAS, gene-based analyses and copy number variation analyses. Community outreach has increased the number of shared datasets from under-represented traits, e.g. cancer, and we continue to contribute to awareness of the lack of population diversity in GWAS. Interoperability of the Catalog has been enhanced through links to other resources including the Polygenic Score Catalog and the International Mouse Phenotyping Consortium, refinements to GWAS trait annotation, and the development of a standard format for GWAS data.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E.et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. - PMC - PubMed
-
- Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E.M., Hercules A., Fumis L., Miranda A., Carvalho-Silva D., Buniello A.et al.. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021; 49:D1311–D1320. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

