Identification of a minority population of LMO2+ breast cancer cells that integrate into the vasculature and initiate metastasis
- PMID: 36351009
- PMCID: PMC10939096
- DOI: 10.1126/sciadv.abm3548
Identification of a minority population of LMO2+ breast cancer cells that integrate into the vasculature and initiate metastasis
Abstract
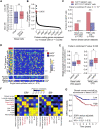
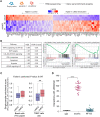
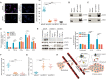
Metastasis is responsible for most breast cancer-related deaths; however, identifying the cellular determinants of metastasis has remained challenging. Here, we identified a minority population of immature THY1+/VEGFA+ tumor epithelial cells in human breast tumor biopsies that display angiogenic features and are marked by the expression of the oncogene, LMO2. Higher abundance of LMO2+ basal cells correlated with tumor endothelial content and predicted poor distant recurrence-free survival in patients. Using MMTV-PyMT/Lmo2CreERT2 mice, we demonstrated that Lmo2 lineage-traced cells integrate into the vasculature and have a higher propensity to metastasize. LMO2 knockdown in human breast tumors reduced lung metastasis by impairing intravasation, leading to a reduced frequency of circulating tumor cells. Mechanistically, we find that LMO2 binds to STAT3 and is required for STAT3 activation by tumor necrosis factor-α and interleukin-6. Collectively, our study identifies a population of metastasis-initiating cells with angiogenic features and establishes the LMO2-STAT3 signaling axis as a therapeutic target in breast cancer metastasis.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

