The memory B cell response to influenza vaccination is impaired in older persons
- PMID: 36351385
- PMCID: PMC9666924
- DOI: 10.1016/j.celrep.2022.111613
The memory B cell response to influenza vaccination is impaired in older persons
Erratum in
-
The memory B cell response to influenza vaccination is impaired in older persons.Cell Rep. 2024 Feb 27;43(2):113745. doi: 10.1016/j.celrep.2024.113745. Epub 2024 Jan 28. Cell Rep. 2024. PMID: 38281131 Free PMC article. No abstract available.
Abstract
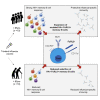
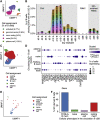
Influenza infection imparts an age-related increase in mortality and morbidity. The most effective countermeasure is vaccination; however, vaccines offer modest protection in older adults. To investigate how aging impacts the memory B cell response, we track hemagglutinin-specific B cells by indexed flow sorting and single-cell RNA sequencing (scRNA-seq) in 20 healthy adults that were administered the trivalent influenza vaccine. We demonstrate age-related skewing in the memory B cell compartment 6 weeks after vaccination, with younger adults developing hemagglutinin-specific memory B cells with an FcRL5+ "atypical" phenotype, showing evidence of somatic hypermutation and positive selection, which happened to a lesser extent in older persons. We use publicly available scRNA-seq from paired human lymph node and blood samples to corroborate that FcRL5+ atypical memory B cells can derive from germinal center (GC) precursors. Together, this study shows that the aged human GC reaction and memory B cell response following vaccination is defective.
Keywords: B cell; CP: Immunology; aging; antibody selection; influenza; memory; vaccination.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Allaire J.J., Xie Y., McPherson J., Luraschi J., Ushey K., Atkins A., Wickham H., Cheng J., Chang W., Iannone R. Dynamic Documents for R; 2021. rmarkdown.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

