Model-based Deep Learning Reconstruction Using a Folded Image Training Strategy for Abdominal 3D T1-weighted Imaging
- PMID: 36351603
- PMCID: PMC10552667
- DOI: 10.2463/mrms.mp.2021-0103
Model-based Deep Learning Reconstruction Using a Folded Image Training Strategy for Abdominal 3D T1-weighted Imaging
Abstract
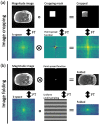
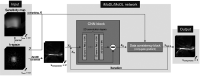
Purpose: To evaluate the feasibility of folded image training strategy (FITS) and the quality of images reconstructed using the improved model-based deep learning (iMoDL) network trained with FITS (FITS-iMoDL) for abdominal MR imaging.
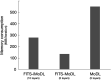
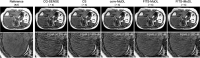
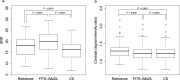
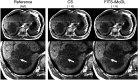
Methods: This retrospective study included abdominal 3D T1-weighted images of 122 patients. In the experimental analyses, peak SNR (PSNR) and structure similarity index (SSIM) of images reconstructed with FITS-iMoDL were compared with those with the following reconstruction methods: conventional model-based deep learning (conv-MoDL), MoDL trained with FITS (FITS-MoDL), total variation regularized compressed sensing (CS), and parallel imaging (CG-SENSE). In the clinical analysis, SNR and image contrast were measured on the reference, FITS-iMoDL, and CS images. Three radiologists evaluated the image quality using a 5-point scale to determine the mean opinion score (MOS).
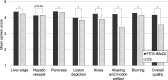
Results: The PSNR of FITS-iMoDL was significantly higher than that of FITS-MoDL, conv-MoDL, CS, and CG-SENSE (P < 0.001). The SSIM of FITS-iMoDL was significantly higher than those of the others (P < 0.001), except for FITS-MoDL (P = 0.056). In the clinical analysis, the SNR of FITS-iMoDL was significantly higher than that of the reference and CS (P < 0.0001). Image contrast was equivalent within an equivalence margin of 10% among these three image sets (P < 0.0001). MOS was significantly improved in FITS-iMoDL (P < 0.001) compared with CS images in terms of liver edge and vessels conspicuity, lesion depiction, artifacts, blurring, and overall image quality.
Conclusion: The proposed method, FITS-iMoDL, allowed a deeper MoDL reconstruction network without increasing memory consumption and improved image quality on abdominal 3D T1-weighted imaging compared with CS images.
Keywords: deep learning; image reconstruction; liver imaging; network training.
Conflict of interest statement
The authors do not have any conflicts of interest.
Figures


References
-
- Lee SM, Lee JM, Ahn SJ, Kang HJ, Yang HK, Yoon JH. LI-RADS version 2017 versus version 2018: Diagnosis of hepatocellular carcinoma on gadoxetate disodium-enhanced MRI. Radiology 2019; 292:655–663. - PubMed
-
- van der Pol CB, Lim CS, Sirlin CB, et al. Accuracy of the liver imaging reporting and data system in computed tomography and magnetic resonance image analysis of hepatocellular carcinoma or overall malignancy—a systematic. Gastroenterology 2019; 156:976–986. - PubMed
-
- Lee S, Kim SS, Roh YH, Choi JY, Park MS, Kim MJ. Diagnostic performance of CT/MRI liver imaging reporting and data system v2017 for hepatocellular carcinoma: A systematic review and meta-analysis. Liver Int 2020; 40:1488–1497. - PubMed
-
- Davenport MS, Viglianti BL, Al-Hawary MM, et al. Comparison of acute transient dyspnea after intravenous administration of gadoxetate disodium and gadobenate dimeglumine: Effect on arterial phase image quality. Radiology 2013; 266:452–461. - PubMed

