Capture at the ER-mitochondrial contacts licenses IP3 receptors to stimulate local Ca2+ transfer and oxidative metabolism
- PMID: 36351901
- PMCID: PMC9646835
- DOI: 10.1038/s41467-022-34365-8
Capture at the ER-mitochondrial contacts licenses IP3 receptors to stimulate local Ca2+ transfer and oxidative metabolism
Abstract
Endoplasmic reticulum-mitochondria contacts (ERMCs) are restructured in response to changes in cell state. While this restructuring has been implicated as a cause or consequence of pathology in numerous systems, the underlying molecular dynamics are poorly understood. Here, we show means to visualize the capture of motile IP3 receptors (IP3Rs) at ERMCs and document the immediate consequences for calcium signaling and metabolism. IP3Rs are of particular interest because their presence provides a scaffold for ERMCs that mediate local calcium signaling, and their function outside of ERMCs depends on their motility. Unexpectedly, in a cell model with little ERMC Ca2+ coupling, IP3Rs captured at mitochondria promptly mediate Ca2+ transfer, stimulating mitochondrial oxidative metabolism. The Ca2+ transfer does not require linkage with a pore-forming protein in the outer mitochondrial membrane. Thus, motile IP3Rs can traffic in and out of ERMCs, and, when 'parked', mediate calcium signal propagation to the mitochondria, creating a dynamic arrangement that supports local communication.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
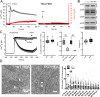

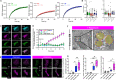

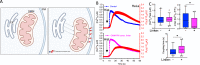
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

