Machine learning for morbid glomerular hypertrophy
- PMID: 36351996
- PMCID: PMC9646707
- DOI: 10.1038/s41598-022-23882-7
Machine learning for morbid glomerular hypertrophy
Abstract
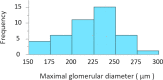
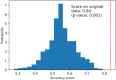
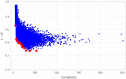
A practical research method integrating data-driven machine learning with conventional model-driven statistics is sought after in medicine. Although glomerular hypertrophy (or a large renal corpuscle) on renal biopsy has pathophysiological implications, it is often misdiagnosed as adaptive/compensatory hypertrophy. Using a generative machine learning method, we aimed to explore the factors associated with a maximal glomerular diameter of ≥ 242.3 μm. Using the frequency-of-usage variable ranking in generative models, we defined the machine learning scores with symbolic regression via genetic programming (SR via GP). We compared important variables selected by SR with those selected by a point-biserial correlation coefficient using multivariable logistic and linear regressions to validate discriminatory ability, goodness-of-fit, and collinearity. Body mass index, complement component C3, serum total protein, arteriolosclerosis, C-reactive protein, and the Oxford E1 score were ranked among the top 10 variables with high machine learning scores using SR via GP, while the estimated glomerular filtration rate was ranked 46 among the 60 variables. In multivariable analyses, the R2 value was higher (0.61 vs. 0.45), and the corrected Akaike Information Criterion value was lower (402.7 vs. 417.2) with variables selected with SR than those selected with point-biserial r. There were two variables with variance inflation factors higher than 5 in those using point-biserial r and none in SR. Data-driven machine learning models may be useful in identifying significant and insignificant correlated factors. Our method may be generalized to other medical research due to the procedural simplicity of using top-ranked variables selected by machine learning.
© 2022. The Author(s).
Conflict of interest statement
Toshio Mochizuki received honoraria for lectures from Otsuka Pharmaceutical Co. Toshio Mochizuki and Hiroshi Kataoka belong to an endowed department sponsored by Otsuka Pharmaceutical Co., Chugai Pharmaceutical Co., Kyowa Hakko Kirin Co., and JMS Co. All other authors have no conflicts of interest to declare.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

