Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid
- PMID: 36352225
- PMCID: PMC9899827
- DOI: 10.1038/s41586-022-05397-3
Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid
Abstract
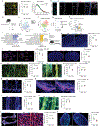
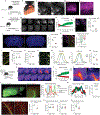
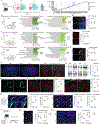
Macrophages are important players in the maintenance of tissue homeostasis1. Perivascular and leptomeningeal macrophages reside near the central nervous system (CNS) parenchyma2, and their role in CNS physiology has not been sufficiently well studied. Given their continuous interaction with the cerebrospinal fluid (CSF) and strategic positioning, we refer to these cells collectively as parenchymal border macrophages (PBMs). Here we demonstrate that PBMs regulate CSF flow dynamics. We identify a subpopulation of PBMs that express high levels of CD163 and LYVE1 (scavenger receptor proteins), closely associated with the brain arterial tree, and show that LYVE1+ PBMs regulate arterial motion that drives CSF flow. Pharmacological or genetic depletion of PBMs led to accumulation of extracellular matrix proteins, obstructing CSF access to perivascular spaces and impairing CNS perfusion and clearance. Ageing-associated alterations in PBMs and impairment of CSF dynamics were restored after intracisternal injection of macrophage colony-stimulating factor. Single-nucleus RNA sequencing data obtained from patients with Alzheimer's disease (AD) and from non-AD individuals point to changes in phagocytosis, endocytosis and interferon-γ signalling on PBMs, pathways that are corroborated in a mouse model of AD. Collectively, our results identify PBMs as new cellular regulators of CSF flow dynamics, which could be targeted pharmacologically to alleviate brain clearance deficits associated with ageing and AD.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests.
J.K. is a scientific advisor and collaborator with PureTech. J.K. and A.D. are holding provisional patent applications related to the findings described herein. R.J.B co-founded and is on the scientific advisory board of C2N Diagnostics. C2N Diagnostics has licensed certain anti-tau antibodies to AbbVie for therapeutic development. He receives research support from Biogen, Eisai, and the DIAN-TU Pharma Consortium. He is also an advisor to Amgen and Hoffman La-Roche.
Figures












Comment in
-
Macrophages bordering the brain parenchyma regulate the flow of cerebrospinal fluid.Nat Rev Immunol. 2023 Jan;23(1):3. doi: 10.1038/s41577-022-00814-0. Nat Rev Immunol. 2023. PMID: 36418433 No abstract available.
-
Macrophages clear the way for CNS fluid flow.Lancet Neurol. 2023 Mar;22(3):194-195. doi: 10.1016/S1474-4422(23)00028-5. Lancet Neurol. 2023. PMID: 36804079 No abstract available.
References
-
- Ginhoux F & Guilliams M Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449 (2016). - PubMed
-
- Masuda T et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748 (2022). - PubMed
-
- Konsman JP, Parnet P & Dantzer R Cytokine-induced sickness behaviour: mechanisms and implications. Trends in Neurosciences 25, 154–159 (2002). - PubMed
Method References:
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG057777/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R37 AG034113/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- P41 GM103422/GM/NIGMS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- UF1 AG032438/AG/NIA NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- U19 AG032438/AG/NIA NIH HHS/United States
- R01 NS092865/NS/NINDS NIH HHS/United States
- R56 AG067764/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- R01 AG062734/AG/NIA NIH HHS/United States
- U01 AG032438/AG/NIA NIH HHS/United States
- R01 AG034113/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P01 AG078106/AG/NIA NIH HHS/United States
- RF1 AG057496/AG/NIA NIH HHS/United States
- R24 GM136766/GM/NIGMS NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

