Locomotion activates PKA through dopamine and adenosine in striatal neurons
- PMID: 36352228
- PMCID: PMC10752255
- DOI: 10.1038/s41586-022-05407-4
Locomotion activates PKA through dopamine and adenosine in striatal neurons
Abstract
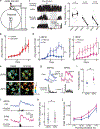
The canonical model of striatal function predicts that animal locomotion is associated with the opposing regulation of protein kinase A (PKA) in direct and indirect pathway striatal spiny projection neurons (SPNs) by dopamine1-7. However, the precise dynamics of PKA in dorsolateral SPNs during locomotion remain to be determined. It is also unclear whether other neuromodulators are involved. Here we show that PKA activity in both types of SPNs is essential for normal locomotion. Using two-photon fluorescence lifetime imaging8-10 of a PKA sensor10 through gradient index lenses, we measured PKA activity within individual SPNs of the mouse dorsolateral striatum during locomotion. Consistent with the canonical view, dopamine activated PKA activity in direct pathway SPNs during locomotion through the dopamine D1 receptor. However, indirect pathway SPNs exhibited a greater increase in PKA activity, which was largely abolished through the blockade of adenosine A2A receptors. In agreement with these results, fibre photometry measurements of an adenosine sensor11 revealed an acute increase in extracellular adenosine during locomotion. Functionally, antagonism of dopamine or adenosine receptors resulted in distinct changes in SPN PKA activity, neuronal activity and locomotion. Together, our results suggest that acute adenosine accumulation interplays with dopamine release to orchestrate PKA activity in SPNs and proper striatal function during animal locomotion.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures

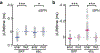


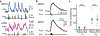


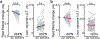




References
-
- Graybiel AM, Aosaki T, Flaherty AW & Kimura M. The basal ganglia and adaptive motor control. Science (80-. ). 265, (1994). - PubMed
-
- Mink JW. The basal ganglia: Focused selection and inhibition of competing motor programs. Prog. Neurobiol. 50, (1996). - PubMed
-
- Klaus A, Alves Da Silva J & Costa RM. What, If, and When to Move: Basal Ganglia Circuits and Self-Paced Action Initiation. Annual Review of Neuroscience vol. 42 (2019). - PubMed
-
- Gerfen CR et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science (80-. ). 250, (1990). - PubMed
Method References
References for Extended Data Figures
-
- Franklin KBJ & Paxinos G The Mouse Brain in Stereotaxic Coordinates (map). Academic Press; (2007).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

