Clinical improvement of DM1 patients reflected by reversal of disease-induced gene expression in blood
- PMID: 36352383
- PMCID: PMC9646470
- DOI: 10.1186/s12916-022-02591-y
Clinical improvement of DM1 patients reflected by reversal of disease-induced gene expression in blood
Abstract
Background: Myotonic dystrophy type 1 (DM1) is an incurable multisystem disease caused by a CTG-repeat expansion in the DM1 protein kinase (DMPK) gene. The OPTIMISTIC clinical trial demonstrated positive and heterogenous effects of cognitive behavioral therapy (CBT) on the capacity for activity and social participations in DM1 patients. Through a process of reverse engineering, this study aims to identify druggable molecular biomarkers associated with the clinical improvement in the OPTIMISTIC cohort.
Methods: Based on full blood samples collected during OPTIMISTIC, we performed paired mRNA sequencing for 27 patients before and after the CBT intervention. Linear mixed effect models were used to identify biomarkers associated with the disease-causing CTG expansion and the mean clinical improvement across all clinical outcome measures.
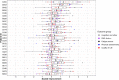
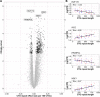
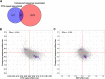
Results: We identified 608 genes for which their expression was significantly associated with the CTG-repeat expansion, as well as 1176 genes significantly associated with the average clinical response towards the intervention. Remarkably, all 97 genes associated with both returned to more normal levels in patients who benefited the most from CBT. This main finding has been replicated based on an external dataset of mRNA data of DM1 patients and controls, singling these genes out as candidate biomarkers for therapy response. Among these candidate genes were DNAJB12, HDAC5, and TRIM8, each belonging to a protein family that is being studied in the context of neurological disorders or muscular dystrophies. Across the different gene sets, gene pathway enrichment analysis revealed disease-relevant impaired signaling in, among others, insulin-, metabolism-, and immune-related pathways. Furthermore, evidence for shared dysregulations with another neuromuscular disease, Duchenne muscular dystrophy, was found, suggesting a partial overlap in blood-based gene dysregulation.
Conclusions: DM1-relevant disease signatures can be identified on a molecular level in peripheral blood, opening new avenues for drug discovery and therapy efficacy assessments.
Keywords: Biomarker; Lifestyle intervention; Myotonic dystrophy type 1; Peripheral blood; RNA-seq; Therapeutic Response.
© 2022. The Author(s).
Conflict of interest statement
R. van Cruchten reports no disclosures relevant to the manuscript; D. van As reports no disclosures relevant to the manuscript; J.C. Glennon reports no disclosures relevant to the manuscript; B. G. M. van Engelen received fees (to the institution) and non-financial support from Fulcrum Therapeutics, Facio Therapies, and Arrowhead Pharmaceuticals during the conduct of the study. In addition, he received grant support from the FP7 European Union grand OPTIMISTIC, Marigold Foundation Canada, Prinses Beartrix Spierfonds, Spieren voor Spieren, FSHD Stichting, and FSHD Society. He also has an unpaid function as the head of the scientific advisory board for Euro-DyMA. P. A. C. ’t Hoen reports no disclosures relevant to the manuscript.
Figures


References
-
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. - DOI - PubMed
-
- Cumming SA, Hamilton MJ, Robb Y, Gregory H, McWilliam C, Cooper A, et al. De novo repeat interruptions are associated with reduced somatic instability and mild or absent clinical features in myotonic dystrophy type 1. Eur J Hum Genet. 2018;26(11):1635–1647. doi: 10.1038/s41431-018-0156-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

