An integrated in silico-in vitro approach for identifying therapeutic targets against osteoarthritis
- PMID: 36352408
- PMCID: PMC9648005
- DOI: 10.1186/s12915-022-01451-8
An integrated in silico-in vitro approach for identifying therapeutic targets against osteoarthritis
Abstract
Background: Without the availability of disease-modifying drugs, there is an unmet therapeutic need for osteoarthritic patients. During osteoarthritis, the homeostasis of articular chondrocytes is dysregulated and a phenotypical transition called hypertrophy occurs, leading to cartilage degeneration. Targeting this phenotypic transition has emerged as a potential therapeutic strategy. Chondrocyte phenotype maintenance and switch are controlled by an intricate network of intracellular factors, each influenced by a myriad of feedback mechanisms, making it challenging to intuitively predict treatment outcomes, while in silico modeling can help unravel that complexity. In this study, we aim to develop a virtual articular chondrocyte to guide experiments in order to rationalize the identification of potential drug targets via screening of combination therapies through computational modeling and simulations.
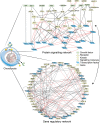
Results: We developed a signal transduction network model using knowledge-based and data-driven (machine learning) modeling technologies. The in silico high-throughput screening of (pairwise) perturbations operated with that network model highlighted conditions potentially affecting the hypertrophic switch. A selection of promising combinations was further tested in a murine cell line and primary human chondrocytes, which notably highlighted a previously unreported synergistic effect between the protein kinase A and the fibroblast growth factor receptor 1.
Conclusions: Here, we provide a virtual articular chondrocyte in the form of a signal transduction interactive knowledge base and of an executable computational model. Our in silico-in vitro strategy opens new routes for developing osteoarthritis targeting therapies by refining the early stages of drug target discovery.
Keywords: Chondrocyte hypertrophy; Computational modeling; Drug targets; In vitro validation; Network of signal transduction; Osteoarthritis; Regulatory network inference; Virtual cell.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures





References
-
- Karsdal MA, Michaelis M, Ladel C, Siebuhr AS, Bihlet AR, Andersen JR, et al. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future. Osteoarthr Cartil. 2016;24:2013–2021. - PubMed
-
- Von Der Mark K, Kirsch T, Nerlich A, Kuss A, Weseloh G, Glückert K, et al. Type x collagen synthesis in human osteoarthritic cartilage. indication of chondrocyte hypertrophy. Arthritis Rheum. 1992;35:806–811. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

