Characterization of an Entner-Doudoroff pathway-activated Escherichia coli
- PMID: 36352474
- PMCID: PMC9648032
- DOI: 10.1186/s13068-022-02219-6
Characterization of an Entner-Doudoroff pathway-activated Escherichia coli
Abstract
Background: Escherichia coli have both the Embden-Meyerhof-Parnas pathway (EMPP) and Entner-Doudoroff pathway (EDP) for glucose breakdown, while the EDP primarily remains inactive for glucose metabolism. However, EDP is a more favorable route than EMPP for the production of certain products.
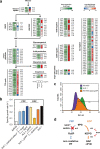
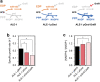
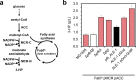
Results: EDP was activated by deleting the pfkAB genes in conjunction with subsequent adaptive laboratory evolution (ALE). The evolved strains acquired mutations in transcriptional regulatory genes for glycolytic process (crp, galR, and gntR) and in glycolysis-related genes (gnd, ptsG, and talB). The genotypic, transcriptomic and phenotypic analyses of those mutations deepen our understanding of their beneficial effects on cellulosic biomass bio-conversion. On top of these scientific understandings, we further engineered the strain to produce higher level of lycopene and 3-hydroxypropionic acid.
Conclusions: These results indicate that the E. coli strain has innate capability to use EDP in lieu of EMPP for glucose metabolism, and this versatility can be harnessed to further engineer E. coli for specific biotechnological applications.
Keywords: 3-Hydroxypropionic acid; Adaptive laboratory evolution; Entner–Doudoroff pathway; Escherichia coli; Glucose metabolism; Phosphofructokinase.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Romano AH, Conway T. Evolution of carbohydrate metabolic pathways. Res Microbiol. 1996;147(6):448–455. - PubMed
-
- Fraenkel DG, Vinopal RT. Carbohydrate metabolism in bacteria. Annu Rev Microbiol. 1973;27(1):69–100.
-
- Ponce E, Garcia M, Munoz ME. Participation of the Entner-Doudoroff pathway in Escherichia coli strains with an inactive phosphotransferase system (PTS – Glc +) in gluconate and glucose batch cultures. Can J Microbiol. 2005;51(11):975–982. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
