Characterization of the complete chloroplast genome of Veronica arvensis and its phylogenomic inference in plantaginaceae
- PMID: 36353054
- PMCID: PMC9639546
- DOI: 10.1080/23802359.2022.2139162
Characterization of the complete chloroplast genome of Veronica arvensis and its phylogenomic inference in plantaginaceae
Abstract
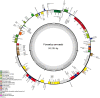
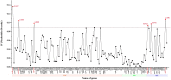
Veronica arvensis, which is an annual flowering plant in the plantain family Plantaginaceae, has commonly used as a Chinese herbal medicine to treat malaria in China. Here, the complete plastome of V. arvensis was successfully assembled based on genome skimming sequencing. The plastome of V. arvensis was 149,386 bp in length, comprising a pair of inverted repeats (IR; 24,946 bp) separated by a large single-copy (LSC) region (82,004 bp) and a small single-copy (SSC) region (17,490 bp). The plastid genome encoded 113 unique genes, consisting of 79 protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Six plastid hotspot regions (trnH-psbA, trnK-rps16, atpI-rps2, ndhF-rpl32, ccsA-ndhD and rps15-ycf1) were identified within Veronica. Phylogenetic analysis showed that the representative species from Veronica was monophyletic. V. persica and V. polita formed a maximum clade, followed by sister to V. arvensis.
Keywords: Plantaginaceae; Veronica arvensis; phylogeny inference; plastid genome.
© 2022 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Albach DC, Meudt HM.. 2010. Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Mol Phylogenet Evol. 54(2):457–471. - PubMed
-
- Baskin J, Baskin C.. 1983. Germination ecology of Veronica arvensis. J Ecol. 71(1):57–68.
-
- Dodsworth S. 2015. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 20(9):525–527. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
