Human Gut Mycobiome and Fungal Community Interaction: The Unknown Musketeer in the Chemotherapy Response Status in Bladder Cancer
- PMID: 36353067
- PMCID: PMC9638757
- DOI: 10.1016/j.euros.2022.06.005
Human Gut Mycobiome and Fungal Community Interaction: The Unknown Musketeer in the Chemotherapy Response Status in Bladder Cancer
Abstract
Background: Until recently, the properties of microbiome and mycobiome in humans and its relevance to disease have largely been unexplored. While the interest of microbiome and malignancy over the past few years have burgeoned with advent of new technologies, no research describing the composition of mycobiome in bladder cancer has been done. Deciphering of the metagenome and its aggregate genetic information can be used to understand the functional properties and relationships between the bacteria, fungi, and cancer.
Objective: The aim of this project is to characterize the compositional range of the normal versus bladder cancer mycobiome of the gut.
Design setting and participants: An internal transcribed spacer (ITS) survey of 52 fecal samples was performed to evaluate the gut mycobiome differences between noncancer controls and bladder cancer patients.
Outcome measurements and statistical analysis: Our study evaluated the differences in mycobiome among patients with bladder cancer, versus matched controls. Our secondary analysis evaluated compositional differences in the gut as a function of response status with neoadjuvant chemotherapy. Data demultiplexing and classification were performed using the QIIME v.1.1.1.1 platform. The Ion Torrent-generated fungal ITS sequence data were processed using QIIME (v.1.9.1), and the reads were demultiplexed, quality filtered, and clustered into operation taxonomic units using default parameters. Alpha and beta diversity were computed and plotted in Phyloseq, principal coordinate analysis was performed on Bray-Curtis dissimilarity indices, and a one-way permutational multivariate analysis of variance was used to test for significant differences between cohorts. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was applied to infer functional categories associated with taxonomic composition.
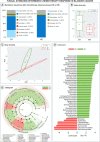
Results and limitations: We found distinctive mycobiome differences between control group (n = 32) and bladder cancer (n = 29) gut flora, and identified an increasing abundance of Tremellales, Hypocreales, and Dothideales. Significant differences in alpha and beta diversity were present between the groups (control vs bladder; p = 0.002), noting distinct compositions within each cohort. A subgroup analysis by sex and neoadjuvant chemotherapy status did not show any further differences in mycobiome composition and diversity. Our results indicate that the gut mycobiome may modulate tumor response to preoperative chemotherapy in bladder cancer patients. We propose that patients with a "favorable" mycobiome composition (eg, high diversity, and low abundance of Agaricomycetes and Saccharomycetes) may have enhanced systemic immune response to chemotherapy through antigen presentation.
Conclusions: Our study is the first to characterize the enteric mycobiome in patients with bladder cancer and describe complex ecological network alterations, indicating complex bacteria-fungi interactions, particularly highlighted among patients with complete neoadjuvant chemotherapy response.
Patient summary: Our study has demonstrated that the composition of stool mycobiome (fungal inhabitants of the gastrointestinal tract) in patients with bladder cancer is different from that in noncancer individuals. Furthermore, when evaluating how patients respond to chemotherapy given prior to their surgery, our study noted significant differences between patients who responded and those who did not.
Keywords: Bladder cancer; Microbiome; Mycobiome; Urothelial cancer.
© 2022 The Authors.
Figures


Comment in
-
The composition and influence of the gut mycobiome in bladder cancer.Nat Rev Urol. 2022 Nov;19(11):634. doi: 10.1038/s41585-022-00668-0. Nat Rev Urol. 2022. PMID: 36224429 No abstract available.
-
Defining the Mycobiome in Bladder Cancer.Eur Urol Open Sci. 2022 Dec 27;48:70-71. doi: 10.1016/j.euros.2022.11.023. eCollection 2023 Feb. Eur Urol Open Sci. 2022. PMID: 36606201 Free PMC article. No abstract available.
References
-
- Sanli O., Dobruch J., Knowles M.A., et al. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

