Development of necroptosis-related gene signature to predict the prognosis of colon adenocarcinoma
- PMID: 36353119
- PMCID: PMC9639779
- DOI: 10.3389/fgene.2022.1051800
Development of necroptosis-related gene signature to predict the prognosis of colon adenocarcinoma
Abstract
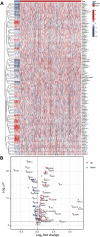
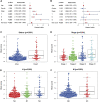
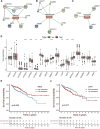
Colon adenocarcinoma (COAD) is a common malignancy and has a high mortality rate. However, the current tumor node metastasis (TNM) staging system is inadequate for prognostic assessment of COAD patients. Therefore, there is an urgent need to identify reliable biomarkers for the prognosis COAD patients. The aberrant expression of necroptosis-related genes (NRGs) is reported to be associated with tumorigenesis and metastasis. In the present work, we compared the expression profiles of NRGs between COAD patients and normal individuals. Based on seven differentially expressed NRGs, a risk score was defined to predict the prognosis of COAD patients. The validation results from both training and independent external cohorts demonstrated that the risk score is able to distinguish the high and low risk COAD patients with higher accuracies, and is independent of the other clinical factors. To facilitate its clinical use, by integrating the proposed risk score, a nomogram was built to predict the risk of individual COAD patients. The C-index of the nomogram is 0.75, indicating the reliability of the nomogram in predicting survival rates. Furthermore, two candidate drugs, namely dapsone and xanthohumol, were screed out and validated by molecular docking, which hold the potential for the treatment of COAD. These results will provide novel clues for the diagnosis and treatment of COAD.
Keywords: colon adenocarcinoma; gene signature; molecular docking; necroptosis; nomogram; survival analysis.
Copyright © 2022 Li, Zhang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification of necroptosis-related genes for predicting prognosis and exploring immune infiltration landscape in colon adenocarcinoma.Front Oncol. 2022 Nov 24;12:941156. doi: 10.3389/fonc.2022.941156. eCollection 2022. Front Oncol. 2022. PMID: 36505813 Free PMC article.
-
Identification and validation of a necroptosis-related gene prognostic signature for colon adenocarcinoma.Transl Cancer Res. 2023 Sep 30;12(9):2239-2255. doi: 10.21037/tcr-23-494. Epub 2023 Aug 31. Transl Cancer Res. 2023. PMID: 37859737 Free PMC article.
-
An Intratumor Heterogeneity-Related Signature for Predicting Prognosis, Immune Landscape, and Chemotherapy Response in Colon Adenocarcinoma.Front Med (Lausanne). 2022 Jul 7;9:925661. doi: 10.3389/fmed.2022.925661. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35872794 Free PMC article.
-
Identification of colon adenocarcinoma necroptosis subtypes and tumor antigens for the development of mRNA vaccines.Heliyon. 2024 Jun 7;10(12):e32531. doi: 10.1016/j.heliyon.2024.e32531. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38952359 Free PMC article.
-
Identification of Transcription Factor-Related Gene Signature and Risk Score Model for Colon Adenocarcinoma.Front Genet. 2021 Sep 17;12:709133. doi: 10.3389/fgene.2021.709133. eCollection 2021. Front Genet. 2021. PMID: 34603375 Free PMC article.
References
LinkOut - more resources
Full Text Sources

