Molecular docking analysis reveals differential binding affinities of multiple classes of selective inhibitors towards cancer-associated KRAS mutants
- PMID: 36353445
- PMCID: PMC9637698
- DOI: 10.1007/s13205-022-03407-9
Molecular docking analysis reveals differential binding affinities of multiple classes of selective inhibitors towards cancer-associated KRAS mutants
Abstract
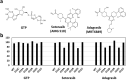
KRAS is the most frequently mutated oncogene in solid cancers, and inhibitors that specifically target the KRAS-G12C mutant were recently approved for clinical use. The limited availability of experimental data pertaining to the sensitivity of KRAS-non-G12C mutants towards RAS inhibitors made it difficult to predict the response of KRAS-mutated cancers towards RAS-targeted therapies. The current study aims at evaluating sensitivity profiles of KRAS-non-G12C mutations towards clinically approved sotorasib and adagrasib, and experimental RAS inhibitors based on binding energies derived through molecular docking analysis. Computationally predicted sensitivities of KRAS mutants conformed with the available but limited experimental data, thus validating the usefulness of molecular docking approach in predicting clinical response towards RAS inhibitor treatment. Our results indicate differential sensitivity of KRAS mutants towards both clinical and experimental therapeutics; while certain mutants exhibited broad cross-resistance to most inhibitors, some mutants showed resistance towards specific inhibitors. These results thus suggest the potential of emergence of more resistance mutations in future towards RAS-targeted therapy and points to an urgent need to develop novel classes of inhibitors that are able to overcome both primary and secondary drug resistance.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03407-9.
Keywords: Adagrasib; Drug sensitivity; KRAS protein; Non-G12C mutations; RAS inhibitors; Sotorasib.
© King Abdulaziz City for Science and Technology 2022.
Conflict of interest statement
Conflict of interestSCM, SA, PSS, VRA, RS, VM, BVLS and RKK declare no potential conflicts of interest.
Figures

References
-
- Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, Johnson ML, Heist RS, Patil T, Riely GJ, Jacobson JO, Yang X, Persky NS, Root DE, Lowder KE, Feng H, Zhang SS, Haigis KM, Hung YP, Sholl LM, Wolpin BM, Wiese J, Christiansen J, Lee J, Schrock AB, Lim LP, Garg K, Li M, Engstrom LD, Waters L, Lawson JD, Olson P, Lito P, Ou SI, Christensen JG, Jänne PA, Aguirre AJ. Acquired resistance to KRASG12C inhibition in cancer. N Engl J Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. - DOI - PMC - PubMed
-
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, Lanman BA, Werner J, Rapaport AS, San Miguel T, Ortiz R, Osgood T, Sun JR, Zhu X, McCarter JD, Volak LP, Houk BE, Fakih MG, O'Neil BH, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Hong DS, Ouyang W, Henary H, Arvedson T, Cee VJ, Lipford JR. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. - DOI - PubMed
-
- Fell JB, Fischer JP, Baer BR, Blake JF, Bouhana K, Briere DM, Brown KD, Burgess LE, Burns AC, Burkard MR, Chiang H, Chicarelli MJ, Cook AW, Gaudino JJ, Hallin J, Hanson L, Hartley DP, Hicken EJ, Hingorani GP, Hinklin RJ, Mejia MJ, Olson P, Otten JN, Rhodes SP, Rodriguez ME, Savechenkov P, Smith DJ, Sudhakar N, Sullivan FX, Tang TP, Vigers GP, Wollenberg L, Christensen JG, Marx MA. Identification of the clinical development candidate MRTX849, a Covalent KRASG12C inhibitor for the treatment of cancer. J Med Chem. 2020;63:6679–6693. doi: 10.1021/acs.jmedchem.9b02052. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

