Transcriptomic analysis of the human placenta reveals trophoblast dysfunction and augmented Wnt signalling associated with spontaneous preterm birth
- PMID: 36353514
- PMCID: PMC9638416
- DOI: 10.3389/fcell.2022.987740
Transcriptomic analysis of the human placenta reveals trophoblast dysfunction and augmented Wnt signalling associated with spontaneous preterm birth
Abstract
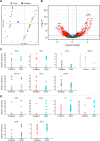
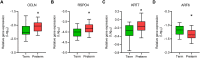
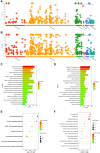
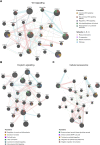
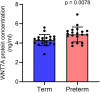
Preterm birth (PTB) is the leading cause of death in under-five children. Worldwide, annually, over 15 million babies are born preterm and 1 million of them die. The triggers and mechanisms of spontaneous PTB remain largely unknown. Most current therapies are ineffective and there is a paucity of reliable predictive biomarkers. Understanding the molecular mechanisms of spontaneous PTB is crucial for developing better diagnostics and therapeutics. To address this need, we conducted RNA-seq transcriptomic analysis, qRT-PCR and ELISA on fresh placental villous tissue from 20 spontaneous preterm and 20 spontaneous term deliveries, to identify genes and signalling pathways involved in the pathogenesis of PTB. Our differential gene expression, gene ontology and pathway analysis revealed several dysregulated genes (including OCLN, OPTN, KRT7, WNT7A, RSPO4, BAMBI, NFATC4, SLC6A13, SLC6A17, SLC26A8 and KLF8) associated with altered trophoblast functions. We identified dysregulated Wnt, oxytocin and cellular senescence signalling pathways in preterm placentas, where augmented Wnt signalling could play a pivotal role in the pathogenesis of PTB due to its diverse biological functions. We also reported two novel targets (ITPR2 and MYLK2) in the oxytocin signalling pathways for further study. Through bioinformatics analysis on DEGs, we identified four key miRNAs, - miR-524-5p, miR-520d-5p, miR-15a-5p and miR-424-5p - which were significantly downregulated in preterm placentas. These miRNAs may have regulatory roles in the aberrant gene expressions that we have observed in preterm placentas. We provide fresh molecular insight into the pathogenesis of spontaneous PTB which may drive further studies to develop new predictive biomarkers and therapeutics.
Keywords: Wnt signalling; miRNAs; placenta; preterm birth; transcriptomic analysis; trophoblast dysfunction.
Copyright © 2022 Akram, Kulkarni, Brook, Wyles and Anumba.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Balda M. S., Whitney J. A., Flores C., González S., Cereijido M., Matter K. (1996). Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 134 (4), 1031–1049. 10.1083/jcb.134.4.1031 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

