A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
- PMID: 36353713
- PMCID: PMC9606447
- DOI: 10.21769/BioProtoc.4535
A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
Abstract
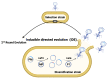
Directed evolution is a powerful technique for identifying beneficial mutations in defined DNA sequences with the goal of improving desired phenotypes. Recent methodological advances have made the evolution of short DNA sequences quick and easy. However, the evolution of DNA sequences >5kb in length, notably gene clusters, is still a challenge for most existing methods. Since many important microbial phenotypes are encoded by multigene pathways, they are usually improved via adaptive laboratory evolution (ALE), which while straightforward to implement can suffer from off-target and hitchhiker mutations that can adversely affect the fitness of the evolved strain. We have therefore developed a new directed evolution method (Inducible Directed Evolution, IDE) that combines the specificity and throughput of recent continuous directed evolution methods with the ease of ALE. Here, we present detailed methods for operating Inducible Directed Evolution (IDE), which enables long (up to 85kb) DNA sequences to be mutated in a high throughput manner via a simple series of incubation steps. In IDE, an intracellular mutagenesis plasmid (MP) tunably mutagenizes the pathway of interest, located on the phagemid (PM). MP contains a mutagenic operon ( danQ926, dam, seqA, emrR, ugi , and cda1 ) that can be expressed via the addition of a chemical inducer. Expression of the mutagenic operon during a cell cycle represses DNA repair mechanisms such as proofreading, translesion synthesis, mismatch repair, and base excision and selection, which leads to a higher mutation rate. Induction of the P1 lytic cycle results in packaging of the mutagenized phagemid, and the pathway-bearing phage particles infect naïve cells, generating a mutant library that can be screened or selected for improved variants. Successive rounds of IDE enable optimization of complex phenotypes encoded by large pathways (as of this writing up to 36 kb), without requiring inefficient transformation steps. Additionally, IDE avoids off-target genomic mutations and enables decoupling of mutagenesis and screening steps, establishing it as a powerful tool for optimizing complex phenotypes in E. coli .
Keywords: Complex Phenotypes; Directed Evolution; Gene Clusters; Inducible Systems; Mutagenesis; P1 Phage; Pathways.
Copyright © 2022 The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interests I.S.A. and N.C. have a patent pending related to this work.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

