The Maize Pathogen Ustilago maydis Secretes Glycoside Hydrolases and Carbohydrate Oxidases Directed toward Components of the Fungal Cell Wall
- PMID: 36354345
- PMCID: PMC9746322
- DOI: 10.1128/aem.01581-22
The Maize Pathogen Ustilago maydis Secretes Glycoside Hydrolases and Carbohydrate Oxidases Directed toward Components of the Fungal Cell Wall
Abstract
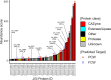
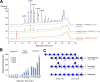
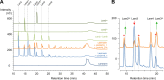
Filamentous fungi are keystone microorganisms in the regulation of many processes occurring on Earth, such as plant biomass decay and pathogenesis as well as symbiotic associations. In many of these processes, fungi secrete carbohydrate-active enzymes (CAZymes) to modify and/or degrade carbohydrates. Ten years ago, while evaluating the potential of a secretome from the maize pathogen Ustilago maydis to supplement lignocellulolytic cocktails, we noticed it contained many unknown or poorly characterized CAZymes. Here, and after reannotation of this data set and detailed phylogenetic analyses, we observed that several CAZymes (including glycoside hydrolases and carbohydrate oxidases) are predicted to act on the fungal cell wall (FCW), notably on β-1,3-glucans. We heterologously produced and biochemically characterized two new CAZymes, called UmGH16_1-A and UmAA3_2-A. We show that UmGH16_1-A displays β-1,3-glucanase activity, with a preference for β-1,3-glucans with short β-1,6 substitutions, and UmAA3_2-A is a dehydrogenase catalyzing the oxidation of β-1,3- and β-1,6-gluco-oligosaccharides into the corresponding aldonic acids. Working on model β-1,3-glucans, we show that the linear oligosaccharide products released by UmGH16_1-A are further oxidized by UmAA3_2-A, bringing to light a putative biocatalytic cascade. Interestingly, analysis of available transcriptomics data indicates that both UmGH16_1-A and UmAA3_2-A are coexpressed, only during early stages of U. maydis infection cycle. Altogether, our results suggest that both enzymes are connected and that additional accessory activities still need to be uncovered to fully understand the biocatalytic cascade at play and its physiological role. IMPORTANCE Filamentous fungi play a central regulatory role on Earth, notably in the global carbon cycle. Regardless of their lifestyle, filamentous fungi need to remodel their own cell wall (mostly composed of polysaccharides) to grow and proliferate. To do so, they must secrete a large arsenal of enzymes, most notably carbohydrate-active enzymes (CAZymes). However, research on fungal CAZymes over past decades has mainly focused on finding efficient plant biomass conversion processes while CAZymes directed at the fungus itself have remained little explored. In the present study, using the maize pathogen Ustilago maydis as model, we set off to evaluate the prevalence of CAZymes directed toward the fungal cell wall during growth of the fungus on plant biomass and characterized two new CAZymes active on fungal cell wall components. Our results suggest the existence of a biocatalytic cascade that remains to be fully understood.
Keywords: CAZymes; Ustilago; beta-glucans; filamentous fungi; fungal cell wall; pathogen; phytopathogens; remodeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lebreton A, Zeng Q, Miyauchi S, Kohler A, Dai YC, Martin FM. 2021. Evolution of the mode of nutrition in symbiotic and saprotrophic fungi in forest Ecosystems. Annu Rev Ecol Evol Syst 52:385–404. 10.1146/annurev-ecolsys-012021-114902. - DOI
-
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Splinter BonDurant S, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavín JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kües U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA 106:1954–1959. 10.1073/pnas.0809575106. - DOI - PMC - PubMed
-
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee Y-H, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kües U, Hibbett DS, Hoffmeister D, Högberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. 10.1126/science.1205411. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

