MicroRNA and mRNA Expression Changes in Glioblastoma Cells Cultivated under Conditions of Neurosphere Formation
- PMID: 36354672
- PMCID: PMC9688839
- DOI: 10.3390/cimb44110360
MicroRNA and mRNA Expression Changes in Glioblastoma Cells Cultivated under Conditions of Neurosphere Formation
Abstract
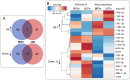
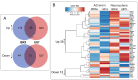
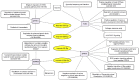
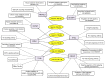
Glioblastoma multiforme (GBM) is one of the most highly metastatic cancers. The study of the pathogenesis of GBM, as well as the development of targeted oncolytic drugs, require the use of actual cell models, in particular, the use of 3D cultures or neurospheres (NS). During the formation of NS, the adaptive molecular landscape of the transcriptome, which includes various regulatory RNAs, changes. The aim of this study was to reveal changes in the expression of microRNAs (miRNAs) and their target mRNAs in GBM cells under conditions of NS formation. Neurospheres were obtained from both immortalized U87 MG and patient-derived BR3 GBM cell cultures. Next generation sequencing analysis of small and long RNAs of adherent and NS cultures of GBM cells was carried out. It was found that the formation of NS proceeds with an increase in the level of seven and a decrease in the level of 11 miRNAs common to U87 MG and BR3, as well as an increase in the level of 38 and a decrease in the level of 12 mRNA/lncRNA. Upregulation of miRNAs hsa-miR: -139-5p; -148a-3p; -192-5p; -218-5p; -34a-5p; and -381-3p are accompanied by decreased levels of their target mRNAs: RTN4, FLNA, SH3BP4, DNPEP, ETS2, MICALL1, and GREM1. Downregulation of hsa-miR: -130b-5p, -25-5p, -335-3p and -339-5p occurs with increased levels of mRNA-targets BDKRB2, SPRY4, ERRFI1 and TGM2. The involvement of SPRY4, ERRFI1, and MICALL1 mRNAs in the regulation of EGFR/FGFR signaling highlights the role of hsa-miR: -130b-5p, -25-5p, -335-3p, and -34a-5p not only in the formation of NS, but also in the regulation of malignant growth and invasion of GBM. Our data provide the basis for the development of new approaches to the diagnosis and treatment of GBM.
Keywords: RNA-seq; cancer stem cells; epithelial to mesenchymal transition; glioblastoma; mRNA; miRNA; neurospheres; pro-neural to mesenchymal transition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

