Ecological and Oceanographic Perspectives in Future Marine Fungal Taxonomy
- PMID: 36354908
- PMCID: PMC9696965
- DOI: 10.3390/jof8111141
Ecological and Oceanographic Perspectives in Future Marine Fungal Taxonomy
Abstract
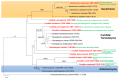
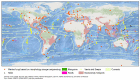
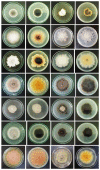
Marine fungi are an ecological rather than a taxonomic group that has been widely researched. Significant progress has been made in documenting their phylogeny, biodiversity, ultrastructure, ecology, physiology, and capacity for degradation of lignocellulosic compounds. This review (concept paper) summarizes the current knowledge of marine fungal diversity and provides an integrated and comprehensive view of their ecological roles in the world's oceans. Novel terms for 'semi marine fungi' and 'marine fungi' are proposed based on the existence of fungi in various oceanic environments. The major maritime currents and upwelling that affect species diversity are discussed. This paper also forecasts under-explored regions with a greater diversity of marine taxa based on oceanic currents. The prospects for marine and semi-marine mycology are highlighted, notably, technological developments in culture-independent sequencing approaches for strengthening our present understanding of marine fungi's ecological roles.
Keywords: culture-independent methods; marine fungal taxonomy; marine-derived fungi; obligate and facultative marine fungi; oceanic currents; upwelling.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments.J Fungi (Basel). 2022 May 8;8(5):491. doi: 10.3390/jof8050491. J Fungi (Basel). 2022. PMID: 35628747 Free PMC article. Review.
-
Biodiversity and Enzyme Activity of Marine Fungi with 28 New Records from the Tropical Coastal Ecosystems in Vietnam.Mycobiology. 2021 Dec 26;49(6):559-581. doi: 10.1080/12298093.2021.2008103. eCollection 2021. Mycobiology. 2021. PMID: 35035248 Free PMC article.
-
Diversity and biogeochemical function of planktonic fungi in the ocean.Prog Mol Subcell Biol. 2012;53:71-88. doi: 10.1007/978-3-642-23342-5_4. Prog Mol Subcell Biol. 2012. PMID: 22222827
-
Biotechnology of marine fungi.Prog Mol Subcell Biol. 2012;53:277-97. doi: 10.1007/978-3-642-23342-5_14. Prog Mol Subcell Biol. 2012. PMID: 22222837 Review.
-
Diversity of fungal isolates from three Hawaiian marine sponges.Microbiol Res. 2009;164(2):233-41. doi: 10.1016/j.micres.2007.07.002. Epub 2007 Aug 6. Microbiol Res. 2009. PMID: 17681460
Cited by
-
OMICS and Other Advanced Technologies in Mycological Applications.J Fungi (Basel). 2023 Jun 19;9(6):688. doi: 10.3390/jof9060688. J Fungi (Basel). 2023. PMID: 37367624 Free PMC article. Review.
-
From marine neglected substrata new fungal taxa of potential biotechnological interest: the case of Pelagia noctiluca.Front Microbiol. 2024 Oct 11;15:1473269. doi: 10.3389/fmicb.2024.1473269. eCollection 2024. Front Microbiol. 2024. PMID: 39464400 Free PMC article.
-
Exploring Fungal Diversity in Seagrass Ecosystems for Pharmaceutical and Ecological Insights.J Fungi (Basel). 2024 Sep 2;10(9):627. doi: 10.3390/jof10090627. J Fungi (Basel). 2024. PMID: 39330387 Free PMC article. Review.
-
Quantification of the dark fungal taxon Cryptomycota using qPCR.Environ Microbiol Rep. 2024 Apr;16(2):e13257. doi: 10.1111/1758-2229.13257. Environ Microbiol Rep. 2024. PMID: 38615691 Free PMC article.
-
Fungal Community Complexity and Stability in Clay Loam and Sandy Soils in Mangrove Ecosystems.J Fungi (Basel). 2025 Mar 28;11(4):262. doi: 10.3390/jof11040262. J Fungi (Basel). 2025. PMID: 40278083 Free PMC article.
References
-
- Hawksworth D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001;105:1422–1432. doi: 10.1017/S0953756201004725. - DOI
-
- Lücking R., Aime M.C., Robbertse B., Miller A.N., Ariyawansa H.A., Aoki T., Cardinali G., Crous P.W., Druzhinina I.S., Geiser D.M., et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 2020;11:14. doi: 10.1186/s43008-020-00033-z. - DOI - PMC - PubMed
-
- Phukhamsakda C., Nilsson R.H., Bhunjun C.S., de Farias A.R.G., Sun Y.R., Wijesinghe S.N., Raza M., Bao D.F., Lu L., Tibpromma S., et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022;114:327–386. doi: 10.1007/s13225-022-00502-3. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

