Applying CRISPR-Cas9 screens to dissect hematological malignancies
- PMID: 36355853
- PMCID: PMC10225491
- DOI: 10.1182/bloodadvances.2022008966
Applying CRISPR-Cas9 screens to dissect hematological malignancies
Abstract
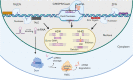
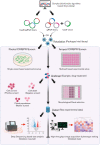
Bit by bit, over the last few decades, functional genomic tools have been piecing together the molecular puzzle driving tumorigenesis in human patients. Nevertheless, our understanding of the role of several genes and regulatory elements that drive critical cancer-associated physiological processes from disease development to progression to spread is very limited, which significantly affects our ability of applying these insights in the context of improved disease management. The recent advent of clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9)-based technology and its application in cancer genomics has, however, allowed the generation of a wealth of knowledge that has helped decipher several critical questions associated with translational cancer research. Precisely, the high-throughput capability coupled with a high level of technological plasticity associated with the CRISPR-Cas9 screens have expanded our horizons from a mere struggle to appreciate cancer as a genetic disease to observing the integrated genomic/epigenomic network of numerous malignancies and correlating it with our present knowledge of drugging strategies to develop innovative approaches for next-generation precision cancer medicine. Specifically, within blood cancers, current CRISPR screens have specifically focused on improving our understanding of drug resistance mechanisms, disease biology, the development of novel therapeutic approaches, and identifying the molecular mechanisms of current therapies, with an underlying aim of improving disease outcomes. Here, we review the development of the CRISPR-Cas9 genome-editing strategy, explicitly focusing on the recent advances in the CRISPR-Cas9-based screening approaches, its current capabilities, limitations, and future applications in the context of hematological malignancies.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
Similar articles
-
Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements.Yi Chuan. 2020 May 20;42(5):435-443. doi: 10.16288/j.yczz.19-390. Yi Chuan. 2020. PMID: 32431295 Review.
-
Current applications and future perspective of CRISPR/Cas9 gene editing in cancer.Mol Cancer. 2022 Feb 21;21(1):57. doi: 10.1186/s12943-022-01518-8. Mol Cancer. 2022. PMID: 35189910 Free PMC article. Review.
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
CRISPR-Cas9, A Promising Therapeutic Tool for Cancer Therapy: A Review.Protein Pept Lett. 2020;27(10):931-944. doi: 10.2174/0929866527666200407112432. Protein Pept Lett. 2020. PMID: 32264803
-
Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing.Drug Deliv Transl Res. 2023 May;13(5):1500-1519. doi: 10.1007/s13346-023-01320-z. Epub 2023 Mar 29. Drug Deliv Transl Res. 2023. PMID: 36988873 Free PMC article. Review.
Cited by
-
Preclinical Anticipation of On- and Off-Target Resistance Mechanisms to Anti-Cancer Drugs: A Systematic Review.Int J Mol Sci. 2024 Jan 5;25(2):705. doi: 10.3390/ijms25020705. Int J Mol Sci. 2024. PMID: 38255778 Free PMC article.
References
-
- Lander ES. The heroes of CRISPR. Cell. 2016;164(1-2):18–28. - PubMed
-
- Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9(3):613–621. - PubMed
-
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (N Y) 2005;151(8):2551–2561. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

