A Review of the Proteomic Profiling of African Viperidae and Elapidae Snake Venoms and Their Antivenom Neutralisation
- PMID: 36355973
- PMCID: PMC9694588
- DOI: 10.3390/toxins14110723
A Review of the Proteomic Profiling of African Viperidae and Elapidae Snake Venoms and Their Antivenom Neutralisation
Abstract
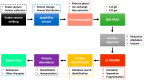
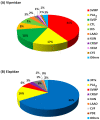
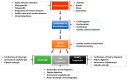
Snakebite envenoming is a neglected tropical disease (NTD) that results from the injection of snake venom of a venomous snake into animals and humans. In Africa (mainly in sub-Saharan Africa), over 100,000 envenomings and over 10,000 deaths per annum from snakebite have been reported. Difficulties in snakebite prevention and antivenom treatment are believed to result from a lack of epidemiological data and underestimated figures on snakebite envenoming-related morbidity and mortality. There are species- and genus-specific variations associated with snake venoms in Africa and across the globe. These variations contribute massively to diverse differences in venom toxicity and pathogenicity that can undermine the efficacy of adopted antivenom therapies used in the treatment of snakebite envenoming. There is a need to profile all snake venom proteins of medically important venomous snakes endemic to Africa. This is anticipated to help in the development of safer and more effective antivenoms for the treatment of snakebite envenoming within the continent. In this review, the proteomes of 34 snake venoms from the most medically important snakes in Africa, namely the Viperidae and Elipdae, were extracted from the literature. The toxin families were grouped into dominant, secondary, minor, and others based on the abundance of the protein families in the venom proteomes. The Viperidae venom proteome was dominated by snake venom metalloproteinases (SVMPs-41%), snake venom serine proteases (SVSPs-16%), and phospholipase A2 (PLA2-17%) protein families, while three-finger toxins (3FTxs-66%) and PLA2s (16%) dominated those of the Elapidae. We further review the neutralisation of these snake venoms by selected antivenoms widely used within the African continent. The profiling of African snake venom proteomes will aid in the development of effective antivenom against snakebite envenoming and, additionally, could possibly reveal therapeutic applications of snake venom proteins.
Keywords: antivenom; elapids; proteomics; snake venom; toxins; venomics; viperids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Defining the pathogenic threat of envenoming by South African shield-nosed and coral snakes (genus Aspidelaps), and revealing the likely efficacy of available antivenom.J Proteomics. 2019 Apr 30;198:186-198. doi: 10.1016/j.jprot.2018.09.019. Epub 2018 Oct 2. J Proteomics. 2019. PMID: 30290233
-
The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms.J Proteomics. 2018 Feb 10;172:173-189. doi: 10.1016/j.jprot.2017.08.016. Epub 2017 Aug 24. J Proteomics. 2018. PMID: 28843532
-
Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom.J Proteomics. 2016 Mar 16;136:262-73. doi: 10.1016/j.jprot.2016.02.006. Epub 2016 Feb 12. J Proteomics. 2016. PMID: 26883873
-
Deciphering toxico-proteomics of Asiatic medically significant venomous snake species: A systematic review and interactive data dashboard.Toxicon. 2024 Nov 6;250:108120. doi: 10.1016/j.toxicon.2024.108120. Epub 2024 Oct 10. Toxicon. 2024. PMID: 39393539
-
Venomous snakes of Costa Rica: biological and medical implications of their venom proteomic profiles analyzed through the strategy of snake venomics.J Proteomics. 2014 Jun 13;105:323-39. doi: 10.1016/j.jprot.2014.02.020. Epub 2014 Feb 24. J Proteomics. 2014. PMID: 24576642 Review.
Cited by
-
Genome assembly and annotation of the Brown-Spotted Pit viper Protobothrops mucrosquamatus.GigaByte. 2023 Nov 7;2023:gigabyte97. doi: 10.46471/gigabyte.97. eCollection 2023. GigaByte. 2023. PMID: 38023064 Free PMC article.
-
Unveiling the functional epitopes of cobra venom cytotoxin by immunoinformatics and epitope-omic analyses.Sci Rep. 2023 Jul 28;13(1):12271. doi: 10.1038/s41598-023-39222-2. Sci Rep. 2023. PMID: 37507457 Free PMC article.
-
Snakebite Management: The Need of Reassessment, International Relations, and Effective Economic Measures to Reduce the Considerable SBE Burden.J Epidemiol Glob Health. 2024 Sep;14(3):586-612. doi: 10.1007/s44197-024-00247-z. Epub 2024 Jun 10. J Epidemiol Glob Health. 2024. PMID: 38856820 Free PMC article. Review.
-
Metabolomics and proteomics: synergistic tools for understanding snake venom inhibition.Arch Toxicol. 2025 Mar;99(3):915-934. doi: 10.1007/s00204-024-03947-4. Epub 2025 Jan 6. Arch Toxicol. 2025. PMID: 39760869 Review.
-
Assessment of the Artemia salina toxicity assay as a substitute of the mouse lethality assay in the determination of venom-induced toxicity and preclinical efficacy of antivenom.Toxicon X. 2024 Apr 3;22:100195. doi: 10.1016/j.toxcx.2024.100195. eCollection 2024 Jun. Toxicon X. 2024. PMID: 38606385 Free PMC article.
References
-
- Kasturiratne A., Wickremasinghe A.R., De Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., De Silva H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:1591–1604. doi: 10.1371/journal.pmed.0050218. - DOI - PMC - PubMed
-
- WHO . Snakebite Envenoming: Strategy for Prevention and Control. World Health Organization; Geneva, Switzerland: 2019. [(accessed on 29 August 2022)]. Available online: https://apps.who.int/iris/handle/10665/312195.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

