First Genome-Based Characterisation and Staphylococcal Enterotoxin Production Ability of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Strains Isolated from Ready-to-Eat Foods in Algiers (Algeria)
- PMID: 36355981
- PMCID: PMC9694651
- DOI: 10.3390/toxins14110731
First Genome-Based Characterisation and Staphylococcal Enterotoxin Production Ability of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Strains Isolated from Ready-to-Eat Foods in Algiers (Algeria)
Abstract
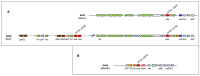
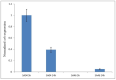
Staphylococcus aureus is a pathogenic microorganism of humans and animals, able to cause foodborne intoxication due to the production of staphylococcal enterotoxins (SEs) and to resist antibiotic treatment as in the case of methicillin-resistant S. aureus (MRSA). In this study, we performed a genomic characterisation of 12 genetically diverse S. aureus strains isolated from ready-to-eat foods in Algiers (Algeria). Moreover, their ability to produce some classical and new staphylococcal enterotoxins (SEs) was investigated. The 12 S. aureus strains resulted to belong to nine known sequence types (STs) and to the novel ST7199 and ST7200. Furthermore, S. aureus SA46 was assigned to the European clone MRSA-ST80-SCCmec-IV. The 12 strains showed a wide endowment of se and sel (staphylococcal enterotoxin-like toxin) genes (sea, seb, sed, seg, seh, sei, selj, sek, sem, sen, seo, seq, ser, selu2, selw, selx, sey, sel30; ψent1-ψent2), including variants and pseudogenes, and harboured the enterotoxin gene cluster (egc) types 1 and 5. Additionally, they produced various amounts of SEA (64.54-345.02 ng/mL), SEB (2871.28-14739.17 ng/mL), SED (322.70-398.94 ng/mL), SEH (not detectable-239.48 ng/mL), and SER (36,720.10-63,176.06 ng/mL) depending on their genotypes. The genetic determinants related to their phenotypic resistance to β-lactams (blaZ, mecA), ofloxacin (gyrA-S84L), erythromycin (ermB), lincomycin (lmrS), kanamycin (aph(3')-III, ant(6)-I), and tetracyclin (tet(L), tet(38)) were also detected. A plethora of virulence-related genes, including major virulence genes such as the tst gene, determinant for the toxic shock syndrome toxin-1, and the lukF-PV and lukS-PV genes, encoding the panton-valentine leukocidin (PVL), were present in the S. aureus strains, highlighting their pathogenic potential. Furthermore, a phylogenomic reconstruction including worldwide foodborne S. aureus showed a clear clustering based on ST and geographical origin rather than the source of isolation.
Keywords: Staphylococcus aureus; antibiotic resistance; food safety; genomics; lukF-PV and lukS-PV genes; methicillin-resistant Staphylococcus aureus (MRSA); panton-valentine leukocidin (PVL); ready-to-eat food; staphylococcal enterotoxins; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

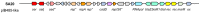

Similar articles
-
Staphylococcal Enterotoxins: Description and Importance in Food.Pathogens. 2024 Aug 9;13(8):676. doi: 10.3390/pathogens13080676. Pathogens. 2024. PMID: 39204276 Free PMC article. Review.
-
Prevalence, Enterotoxigenic Potential and Antimicrobial Resistance of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Algerian Ready to Eat Foods.Toxins (Basel). 2021 Nov 24;13(12):835. doi: 10.3390/toxins13120835. Toxins (Basel). 2021. PMID: 34941673 Free PMC article.
-
Phenotypic and genotypic characterization of PVL-positive Staphylococcus aureus isolated from retail foods in China.Int J Food Microbiol. 2019 Sep 2;304:119-126. doi: 10.1016/j.ijfoodmicro.2019.05.021. Epub 2019 May 27. Int J Food Microbiol. 2019. PMID: 31195259
-
Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci from Japanese retail ready-to-eat raw fish.Int J Food Microbiol. 2012 Jun 1;156(3):286-9. doi: 10.1016/j.ijfoodmicro.2012.03.022. Epub 2012 Mar 28. Int J Food Microbiol. 2012. PMID: 22541390
-
Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen.Lett Appl Microbiol. 2017 Jun;64(6):409-418. doi: 10.1111/lam.12735. Epub 2017 May 3. Lett Appl Microbiol. 2017. PMID: 28304109 Review.
Cited by
-
Staphylococcal Enterotoxins: Description and Importance in Food.Pathogens. 2024 Aug 9;13(8):676. doi: 10.3390/pathogens13080676. Pathogens. 2024. PMID: 39204276 Free PMC article. Review.
-
Molecular Epidemiology and Characterization of Multidrug-Resistant MRSA ST398 and ST239 in Himachal Pradesh, India.Infect Drug Resist. 2023 Apr 20;16:2339-2348. doi: 10.2147/IDR.S409037. eCollection 2023. Infect Drug Resist. 2023. PMID: 37125211 Free PMC article.
-
A Novel Approach Based on Real-Time PCR with High-Resolution Melting Analysis for the Simultaneous Identification of Staphylococcus aureus and Staphylococcus argenteus.Foods. 2024 Sep 22;13(18):3004. doi: 10.3390/foods13183004. Foods. 2024. PMID: 39335932 Free PMC article.
-
A Comprehensive Review of Detection Methods for Staphylococcus aureus and Its Enterotoxins in Food: From Traditional to Emerging Technologies.Toxins (Basel). 2025 Jun 23;17(7):319. doi: 10.3390/toxins17070319. Toxins (Basel). 2025. PMID: 40711131 Free PMC article. Review.
-
Exploring diflunisal as a synergistic agent against Staphylococcus aureus biofilm formation.Front Microbiol. 2024 Sep 25;15:1399996. doi: 10.3389/fmicb.2024.1399996. eCollection 2024. Front Microbiol. 2024. PMID: 39386371 Free PMC article.
References
-
- Dicks J., Turnbull J.D., Russell J., Parkhill J., Alexander S. Genome sequencing of a historic Staphylococcus aureus collection reveals new enterotoxin genes and sheds light on the evolution and genomic organization of this key virulence gene family. J. Bacteriol. 2021;203:e00587-20. doi: 10.1128/JB.00587-20. - DOI - PMC - PubMed
-
- Mekhloufi O.A., Chieffi D., Hammoudi A., Bensefia S.A., Fanelli F., Fusco V. Prevalence, enterotoxigenic potential and antimicrobial resistance of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) isolated from Algerian ready to eat foods. Toxins. 2021;13:835. doi: 10.3390/toxins13120835. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

