Immunological signature in human cases of monkeypox infection in 2022 outbreak: an observational study
- PMID: 36356606
- PMCID: PMC9761565
- DOI: 10.1016/S1473-3099(22)00662-4
Immunological signature in human cases of monkeypox infection in 2022 outbreak: an observational study
Abstract
Background: An unprecedented global monkeypox outbreak started in May, 2022. No data are yet available about the dynamics of the immune response against monkeypox virus. The aim of this study was to describe kinetics of T-cell response, inflammatory profile, and pox-specific T-cell induction in patients with laboratory-confirmed monkeypox.
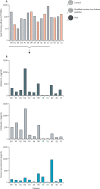
Methods: 17 patients with laboratory-confirmed monkeypox admitted at the Lazzaro Spallanzani National Institute for Infectious Diseases (Rome, Italy), from May 19, to July 7, 2022, were tested for differentiation and activation profile of CD4 and CD8 T (expression of CD38, PD-1, and CD57 assessed by flow cytometry), frequency of pox-specific T cells (by standard interferon-γ ELISpot), and release of interleukin (IL)-1β, IL-6, IL-8, and tumour necrosis factor (TNF) in plasma (by ELISA). All patients were tested 10-12 days after symptoms onset. In a subgroup of nine patients with a laboratory-confirmed monkeypox, the kinetics of the immune response were analysed longitudinally according to timing from symptoms onset and compared with ten healthy donors (ie, health-care workers recruited from the same institution).
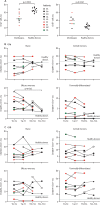
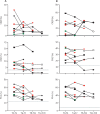
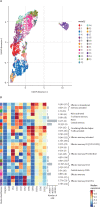
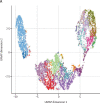
Findings: Among the 17 patients, ten were HIV negative and seven HIV positive, all with good viro-immunological status. On days 0-3 from symptom onset, patients with laboratory-confirmed monkeypox were characterised by a statistically significant reduction in CD4+ T cells (p=0·0011) and a concurrent increase of CD8+ T cells (p=0·0057) compared with healthy donors. A lower proportion of naive (CD45RA+CD27+) CD4+ T cells was observed in six (67%) of nine patients and a concomitant higher proportion of effector memory (CD45RA-CD27-) CD4+ T cells in all patients; this skewed immune profile tended to normalise over time. A similar differentiated profile was also observed in CD8+ T cells with a consistent expansion of terminally differentiated CD8+ T cells. Patients with monkeypox had a higher proportion of CD4+CD38+ and CD38+CD8+ T-cells than healthy donors, which normalised after 12-20 days from symptom onset. The expression of PD-1 and CD57 on CD4+ and CD8+ T-cells showed kinetics similar to that observed for CD38. Furthermore, the inflammatory cytokines (IL-1β, IL-6, IL-8, and TNF) were higher in patients with monkeypox than in healthy donors and, although they decreased over time, they remained elevated after recovery. Almost all patients (15 [94%] of 16) developed a pox-specific Th1 response. No differences in immune cells profile were observed between patients with and without HIV, whereas paucysimptomatic patients (without systemic symptoms, with less than five skin lesions, and no other mucosal localisation of monkeypox) showed a less perturbed immune profile early after symptom onset.
Interpretation: Our data showed the immunological signature of monkeypox virus infection, characterised by an early expansion of activated effector CD4+ and CD8+ T cells that persisted over time. Almost all patients, even regardless of HIV infection, developed a poxvirus-specific Th1 cell response. These results might have implications on the expected immunogenicity of monkeypox vaccination, suggesting that it might not be necessary to vaccinate people who have already been infected.
Funding: Italian Ministry of Health.
Translation: For the Italian translation of the abstract see Supplementary Materials section.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures


References
-
- Centers for Disease Control. 2022 monkeypox outbreak global map. Sept 20, 2022 (accessed Sept 21, 2022).
-
- Thornhill JP, Barkati S, Walmsley S, et al. Monkeypox virus infection in humans across 16 countries-April–June 2022. N Engl J Med. 2022;387:679–691. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

