Long-term SARS-CoV-2 surveillance in the wastewater of Stockholm: What lessons can be learned from the Swedish perspective?
- PMID: 36356735
- PMCID: PMC9640212
- DOI: 10.1016/j.scitotenv.2022.160023
Long-term SARS-CoV-2 surveillance in the wastewater of Stockholm: What lessons can be learned from the Swedish perspective?
Abstract
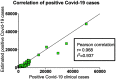
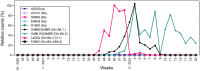
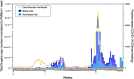
Wastewater-based epidemiology (WBE) can be used to track the spread of SARS-CoV-2 in a population. This study presents the learning outcomes from over two-year long monitoring of SARS-CoV-2 in Stockholm, Sweden. The three main wastewater treatment plants in Stockholm, with a total of six inlets, were monitored from April 2020 until June 2022 (in total 600 samples). This spans five major SARS-CoV-2 waves, where WBE data provided early warning signals for each wave. Further, the measured SARS-CoV-2 content in the wastewater correlated significantly with the level of positive COVID-19 tests (r = 0.86; p << 0.0001) measured by widespread testing of the population. Moreover, as a proof-of-concept, six SARS-CoV-2 variants of concern were monitored using hpPCR assay, demonstrating that variants can be traced through wastewater monitoring. During this long-term surveillance, two sampling protocols, two RNA concentration/extraction methods, two calculation approaches, and normalization to the RNA virus Pepper mild mottle virus (PMMoV) were evaluated. In addition, a study of storage conditions was performed, demonstrating that the decay of viral RNA was significantly reduced upon the addition of glycerol to the wastewater before storage at -80 °C. Our results provide valuable information that can facilitate the incorporation of WBE as a prediction tool for possible future outbreaks of SARS-CoV-2 and preparations for future pandemics.
Keywords: COVID-19; Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); Sewage surveillance; Storage conditions; Wastewater-based epidemiology (WBE); hpPCR.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.v., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/J.ENVRES.2020.110092. - DOI - PMC - PubMed
-
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

