Investigation of in vitro histone H3 glycosylation using H3 tail peptides
- PMID: 36357422
- PMCID: PMC9649660
- DOI: 10.1038/s41598-022-21883-0
Investigation of in vitro histone H3 glycosylation using H3 tail peptides
Abstract
Posttranslational modifications (PTMs) on histone tails regulate eukaryotic gene expression by impacting the chromatin structure and by modulating interactions with other cellular proteins. One such PTM has been identified as serine and threonine glycosylation, the introduction of the ß-N-acetylglucosamine (GlcNAc) moiety on histone H3 tail at position Ser10 and Thr32. The addition of the ß-O-GlcNAc moiety on serine or threonine residues is facilitated by the O-GlcNAc transferase (OGT), and can be removed by the action of O-GlcNAcase (OGA). Conflicting reports on histone tail GlcNAc modification in vivo prompted us to investigate whether synthetic histone H3 tail peptides in conjunction with other PTMs are substrates for OGT and OGA in vitro. Our enzymatic assays with recombinantly expressed human OGT revealed that the unmodified and PTM-modified histone H3 tails are not substrates for OGT at both sites, Ser10 and Thr32. In addition, full length histone H3 was not a substrate for OGT. Conversely, our work demonstrates that synthetic peptides containing the GlcNAc functionality at Ser10 are substrates for recombinantly expressed human OGA, yielding deglycosylated histone H3 peptides. We also show that the catalytic domains of human histone lysine methyltransferases G9a, GLP and SETD7 and histone lysine acetyltransferases PCAF and GCN5 do somewhat tolerate glycosylated H3Ser10 close to lysine residues that undergo methylation and acetylation reactions, respectively. Overall, this work indicates that GlcNAcylation of histone H3 tail peptide in the presence of OGT does not occur in vitro.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

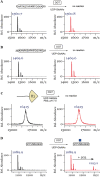

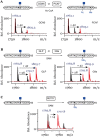
References
-
- Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

