A new catalytic site functioning in antigen cleavage by H34 catalytic antibody light chain
- PMID: 36357546
- PMCID: PMC9649737
- DOI: 10.1038/s41598-022-23689-6
A new catalytic site functioning in antigen cleavage by H34 catalytic antibody light chain
Abstract
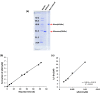
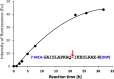
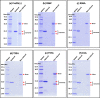
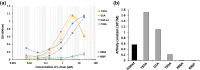
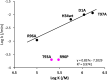
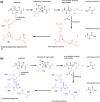
The cleavage reactions of catalytic antibodies are mediated by a serine protease mechanism involving a catalytic triad composed of His, Ser, and Asp residues, which reside in the variable region. Recently, we discovered a catalytic antibody, H34 wild type (H34wt), that is capable of enzymatically cleaving an immune-check point PD-1 peptide and recombinant PD-1; however, H34wt does not contain His residues in the variable region. To clarify the reason behind the catalytic features of H34wt and the amino acid residues involved in the catalytic reaction, we performed site-directed mutagenesis focusing on the amino acid residues involved in the cleavage reaction, followed by catalytic activity tests, immunological reactivity evaluation, and molecular modeling. The results revealed that the cleavage reaction by H34wt proceeds through the action of a new catalytic site composed of Arg, Thr, and Gln. This new scheme differs from that of the serine protease mechanism of catalytic antibodies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Paul S, et al. Catalytic hydrolysis of vasoactive intestinal peptide by human autoantibody. Science. 1989;244:1158–1162. - PubMed
-
- Hifumi E, Kondo H, Mitsuda Y, Uda T. Catalytic features of monoclonal antibody i41SL1-2 subunits. Biotechnol. Bioeng. 2003;84:485–493. - PubMed
-
- Mei S, Mody B, Eklund SH, Paul S. Vasoactive intestinal peptide hydrolysis by antibody light chains (Communication) J. Biol. Chem. 1991;266:15571–15574. - PubMed
-
- Gao QS, et al. Molecular cloning of a proteolytic antibody light chain. J. Biol. Chem. 1994;269:32389–32393. - PubMed
-
- Taguchi H, et al. Autoantibody-catalyzed hydrolysis of Amyloid β peptide. J. Biol. Chem. 2008;283:4714–4722. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

