Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer
- PMID: 36357680
- PMCID: PMC10338177
- DOI: 10.1038/s41591-022-02047-z
Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer
Abstract
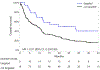
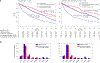
Circulating tumor DNA (ctDNA) sequencing guides therapy decisions but has been studied mostly in small cohorts without sufficient follow-up to determine its influence on overall survival. We prospectively followed an international cohort of 1,127 patients with non-small-cell lung cancer and ctDNA-guided therapy. ctDNA detection was associated with shorter survival (hazard ratio (HR), 2.05; 95% confidence interval (CI), 1.74-2.42; P < 0.001) independently of clinicopathologic features and metabolic tumor volume. Among the 722 (64%) patients with detectable ctDNA, 255 (23%) matched to targeted therapy by ctDNA sequencing had longer survival than those not treated with targeted therapy (HR, 0.63; 95% CI, 0.52-0.76; P < 0.001). Genomic alterations in ctDNA not detected by time-matched tissue sequencing were found in 25% of the patients. These ctDNA-only alterations disproportionately featured subclonal drivers of resistance, including RICTOR and PIK3CA alterations, and were associated with short survival. Minimally invasive ctDNA profiling can identify heterogeneous drivers not captured in tissue sequencing and expand community access to life-prolonging therapy.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
J.J. has a patent licensed by MDSeq. A.N. reports serving as a one-time paid consultant for Bayer. P.K.P. receives compensation for consulting or advisory board participation from Bicara Therapeutics, Boehringer Ingelheim, EMD Serono, GlaxoSmithKline, Takeda Pharmaceuticals, WC Communications and Xencor and receives honoraria for participation in CME educational programs from PeerVoice, ACE Oncology Research to Practice, Clinical Care Options, Spring to Life and Touch Independent Medical Education. J.E.C. has served as a consultant for Astra Zeneca, Bristol-Myers Squib, Genentech, Merck, Flame Biosciences, Novartis, Regeneron-Sanofi, Guardant Health and Janssen and has received research funding to her institution from Astra Zeneca, Bristol-Myers Squib, Genentech and Merck. A.R.B. has stock ownership in Johnson & Johnson. R.B. reports a grant from Archer, honoraria for advisory board participation from Loxo oncology and speaking fees from Illumina. A.Z. has received speaking fees from Illumina. D.C. has consulted with/received honoraria from Pfizer, Loxo/Lilly Oncology, BridgeBio, FORE Therapeutics, Scorpion Therapeutics and Vividion Therapeutics. Y.R.M.-G. acknowledges receipt of training through an institutional K30 grant from the NIH (CTSA UL1TR00457). She has received funding from a Kristina M. Day Young Investigator Award from Conquer Cancer, the ASCO Foundation, funded by Charles M. Baum and Carol A. Baum. She reports travel, accommodation and expenses from AstraZeneca and honoraria from Virology Education. She acknowledges research funding to the institution from Loxo Oncology at Eli Lilly, Elucida Oncology, Taiho Oncology, Hengrui USA/Jiangsu Hengrui Pharmaceuticals and Endeavor Biomedicines. She acknowledges royalties from Rutgers University Press and Wolters Kluwer. H.-Y.T. has received academic travel support from Resolution Bioscience. C.X. has received honoraria from AstraZeneca, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Lilly, Merck & Co., Novartis, Pfizer and Roche; has received research support from BeiGene and Hengrui Pharmaceutical; and has received reimbursement for travel and accommodation expenses from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Lilly, Merck & Co., Novartis, Pfizer and Roche. B.D. reports equity interest in Eli Lilly and Company, Roche and CVS Health and family member involvement with Eli Lilly and Company and CVS Health. H.A.Y. has consulted for AstraZeneca, Blueprint Medicine, Janssen Oncology, Cullinan and Daiichi. Her institution has received research funding for clinical trials from AstraZeneca, Daiichi, Pfizer, Novartis, Cullinan Oncology and Lilly. M.O. reports personal fees from PharmaMar, personal fees from Novartis, personal fees from Targeted Oncology, personal fees from Bristol-Myers Squibb, personal fees from Merck Sharp & Dohme, personal fees from Jazz Pharmaceuticals and personal fees from Astro Pharmaceuticals, outside the submitted work. M.D.H. reports grants from BMS and personal fees from Achilles, Adagene, Adicet, Arcus, AstraZeneca, Blueprint, BMS, DaVolterra, Eli Lilly, Genentech/Roche, Genzyme/Sanofi, Janssen, Immunai, Instil Bio, Mana Therapeutics, Merck, Mirati, Natera, Pact Pharma, Shattuck Labs and Regeneron, as well as equity options from Factorial, Immunai, Shattuck Labs and Arcus. A patent filed by Memorial Sloan Kettering related to the use of TMB to predict response to immunotherapy (PCT/US2015/062208) is pending and licensed by PGDx. P.L. is listed as an inventor on patent applications filed by MSKCC that describe approaches to treat
Figures






Comment in
-
Circulating tumor DNA as a novel prognostic indicator.Nat Med. 2022 Nov;28(11):2255-2256. doi: 10.1038/s41591-022-02068-8. Nat Med. 2022. PMID: 36357679 No abstract available.
-
New data confirm clinical utility of ctDNA.Nat Rev Clin Oncol. 2023 Feb;20(2):63. doi: 10.1038/s41571-022-00716-z. Nat Rev Clin Oncol. 2023. PMID: 36473992 No abstract available.
References
-
- Pasche B & Grant SC Non-small cell lung cancer and precision medicine a model for the incorporation of genomic features into clinical trial design. JAMA 311, 1975 (2014). - PubMed
-
- Bruno DS et al. Racial disparities in biomarker testing and clinical trial enrollment in non-small cell lung cancer (NSCLC). J. Clin. Oncol 39, 9005 (2021).
-
- Robert NJ et al. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J. Clin. Oncol. 39, 9004 (2021).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

