Immune correlates analysis of the ENSEMBLE single Ad26.COV2.S dose vaccine efficacy clinical trial
- PMID: 36357712
- PMCID: PMC10166187
- DOI: 10.1038/s41564-022-01262-1
Immune correlates analysis of the ENSEMBLE single Ad26.COV2.S dose vaccine efficacy clinical trial
Abstract
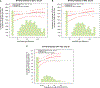
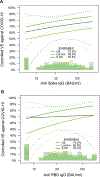
Measuring immune correlates of disease acquisition and protection in the context of a clinical trial is a prerequisite for improved vaccine design. We analysed binding and neutralizing antibody measurements 4 weeks post vaccination as correlates of risk of moderate to severe-critical COVID-19 through 83 d post vaccination in the phase 3, double-blind placebo-controlled phase of ENSEMBLE, an international randomized efficacy trial of a single dose of Ad26.COV2.S. We also evaluated correlates of protection in the trial cohort. Of the three antibody immune markers we measured, we found most support for 50% inhibitory dilution (ID50) neutralizing antibody titre as a correlate of risk and of protection. The outcome hazard ratio was 0.49 (95% confidence interval 0.29, 0.81; P = 0.006) per 10-fold increase in ID50; vaccine efficacy was 60% (43%, 72%) at non-quantifiable ID50 (<2.7 IU50 ml-1) and increased to 89% (78%, 96%) at ID50 = 96.3 IU50 ml-1. Comparison of the vaccine efficacy by ID50 titre curves for ENSEMBLE-US, the COVE trial of the mRNA-1273 vaccine and the COV002-UK trial of the AZD1222 vaccine supported the ID50 titre as a correlate of protection across trials and vaccine types.
Trial registration: ClinicalTrials.gov NCT04505722.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures




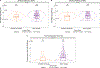
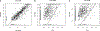





Update of
-
Immune Correlates Analysis of a Single Ad26.COV2.S Dose in the ENSEMBLE COVID-19 Vaccine Efficacy Clinical Trial.medRxiv [Preprint]. 2022 Apr 12:2022.04.06.22272763. doi: 10.1101/2022.04.06.22272763. medRxiv. 2022. Update in: Nat Microbiol. 2022 Dec;7(12):1996-2010. doi: 10.1038/s41564-022-01262-1. PMID: 35441174 Free PMC article. Updated. Preprint.
References
-
- US Food and Drug Administration. Janssen COVID-19 Vaccine. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-dise... Last updated 19 Jan, 2022. Access date 23 Jan, 2022.
-
- World Health Organization. COVID-19 vaccines WHO EUL issued. https://extranet.who.int/pqweb/vaccines/vaccinescovid-19-vaccine-eul-issued Access date 24 Jan, 2022.
-
- European Commission. European Commission authorises fourth safe and effective vaccine against COVID-19. Press release. 11 March 2021. https://ec.europa.eu/commission/presscorner/detail/en/ip_21_1085 Access date 24 Jan, 2022.

