Genomic trajectories of a near-extinction event in the Chatham Island black robin
- PMID: 36357860
- PMCID: PMC9647977
- DOI: 10.1186/s12864-022-08963-1
Genomic trajectories of a near-extinction event in the Chatham Island black robin
Abstract
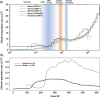
Background: Understanding the micro--evolutionary response of populations to demographic declines is a major goal in evolutionary and conservation biology. In small populations, genetic drift can lead to an accumulation of deleterious mutations, which will increase the risk of extinction. However, demographic recovery can still occur after extreme declines, suggesting that natural selection may purge deleterious mutations, even in extremely small populations. The Chatham Island black robin (Petroica traversi) is arguably the most inbred bird species in the world. It avoided imminent extinction in the early 1980s and after a remarkable recovery from a single pair, a second population was established and the two extant populations have evolved in complete isolation since then. Here, we analysed 52 modern and historical genomes to examine the genomic consequences of this extreme bottleneck and the subsequent translocation.
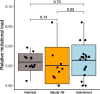
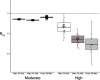
Results: We found evidence for two-fold decline in heterozygosity and three- to four-fold increase in inbreeding in modern genomes. Moreover, there was partial support for temporal reduction in total load for detrimental variation. In contrast, compared to historical genomes, modern genomes showed a significantly higher realised load, reflecting the temporal increase in inbreeding. Furthermore, the translocation induced only small changes in the frequency of deleterious alleles, with the majority of detrimental variation being shared between the two populations.
Conclusion: Our results highlight the dynamics of mutational load in a species that recovered from the brink of extinction, and show rather limited temporal changes in mutational load. We hypothesise that ancestral purging may have been facilitated by population fragmentation and isolation on several islands for thousands of generations and may have already reduced much of the highly deleterious load well before human arrival and introduction of pests to the archipelago. The majority of fixed deleterious variation was shared between the modern populations, but translocation of individuals with low mutational load could possibly mitigate further fixation of high-frequency deleterious variation.
Keywords: Bottleneck; Genomics; Inbreeding; Mutational load; Near-extinction; Translocation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Severe inbreeding depression and no evidence of purging in an extremely inbred wild species--the Chatham Island black robin.Evolution. 2014 Apr;68(4):987-95. doi: 10.1111/evo.12315. Epub 2013 Dec 4. Evolution. 2014. PMID: 24303793
-
Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae).Mol Ecol. 2004 Dec;13(12):3709-21. doi: 10.1111/j.1365-294X.2004.02368.x. Mol Ecol. 2004. PMID: 15548285
-
From high masked to high realized genetic load in inbred Scandinavian wolves.Mol Ecol. 2023 Apr;32(7):1567-1580. doi: 10.1111/mec.16802. Epub 2022 Dec 13. Mol Ecol. 2023. PMID: 36458895
-
Purging and accumulation of genetic load in conservation.Trends Ecol Evol. 2023 Oct;38(10):961-969. doi: 10.1016/j.tree.2023.05.008. Epub 2023 Jun 19. Trends Ecol Evol. 2023. PMID: 37344276 Review.
-
Deleterious Variation in Natural Populations and Implications for Conservation Genetics.Annu Rev Anim Biosci. 2023 Feb 15;11:93-114. doi: 10.1146/annurev-animal-080522-093311. Epub 2022 Nov 4. Annu Rev Anim Biosci. 2023. PMID: 36332644 Free PMC article. Review.
Cited by
-
Genetic Load and Adaptive Potential of a Recovered Avian Species that Narrowly Avoided Extinction.Mol Biol Evol. 2023 Dec 1;40(12):msad256. doi: 10.1093/molbev/msad256. Mol Biol Evol. 2023. PMID: 37995319 Free PMC article.
-
Genetic purging in an island-endemic pigeon recovering from the brink of extinction.Commun Biol. 2025 Jul 15;8(1):1051. doi: 10.1038/s42003-025-08476-z. Commun Biol. 2025. PMID: 40670606 Free PMC article.
-
Biocultural aspects of species extinctions.Camb Prism Extinct. 2023 Aug 16;1:e22. doi: 10.1017/ext.2023.20. eCollection 2023. Camb Prism Extinct. 2023. PMID: 40078689 Free PMC article. Review.
-
Range-wide and temporal genomic analyses reveal the consequences of near-extinction in Swedish moose.Commun Biol. 2023 Oct 17;6(1):1035. doi: 10.1038/s42003-023-05385-x. Commun Biol. 2023. PMID: 37848497 Free PMC article.
-
Adaptation to the High-Arctic island environment despite long-term reduced genetic variation in Svalbard reindeer.iScience. 2023 Sep 3;26(10):107811. doi: 10.1016/j.isci.2023.107811. eCollection 2023 Oct 20. iScience. 2023. PMID: 37744038 Free PMC article.
References
-
- Madsen T, Shine R, Olsson M, Wittzell H. Restoration of an inbred adder population. Nature. 1999;402:34–35.
-
- White KL, Eason DK, Jamieson IG, Robertson BC. Evidence of inbreeding depression in the critically endangered parrot, the kakapo. Anim Conserv. 2015;18:341–347.
-
- Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am Nat. 1995;146:489–518.
-
- van Oosterhout C. Mutation load is the spectre of species conservation. Nat Ecol Evol. 2020;4:1004–1006. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

